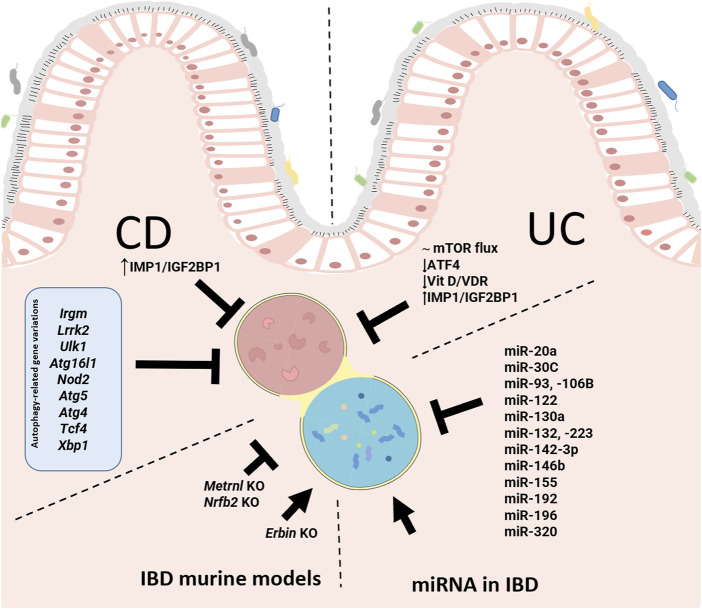
FIGURE 2.
Pathways and genetic and epigenetic basis of autophagy involved in the pathophysiology of inflammatory bowel disease (IBD). In Crohn’s Disease (CD), the mutation or deletion of some autophagy-related genes (blue box) has been related with CD. IGF2BP1/IMP1 has been implicated in autophagy inhibition in CD and Ulcerative Colitis (UC). In UC patients, there is a reduction of the autophagy process through the reduction of ATF4 activity, mTOR-dependent autophagy flux deficiency and low VDR expression and defective vitamin D/VDR signalling. The illustration also features miRNAs described as modulators of autophagy in IBD patients. Finally, we can see the advances made with murine models of IBD that link autophagy to IBD pathology. IGF2BP1/IMP1, Insulin-like growth factor 2 RNA binding protein 1; IRGM, immunity-related GTPase M; LRRK2, leucine-rich repeat serine/threonine-protein kinase 2; ULK-1, Unc-51-like kinase-1; ATG16L1, autophagy-related protein 16-like protein 1; NOD2, nucleotide binding oligomerization domain containing protein 2; ATG5, autophagy-related gen 5; ATG4, autophagy-related gen 4; TCF4, transcription factor 4; XBP1, X box-binding protein 1 spliced-1; mTOR, mammalian target of rapamycin; ATF4, autophagy-related transcription factor 4; VDR, vitamin D receptor; NRBF2, nuclear receptor binding factor 2.