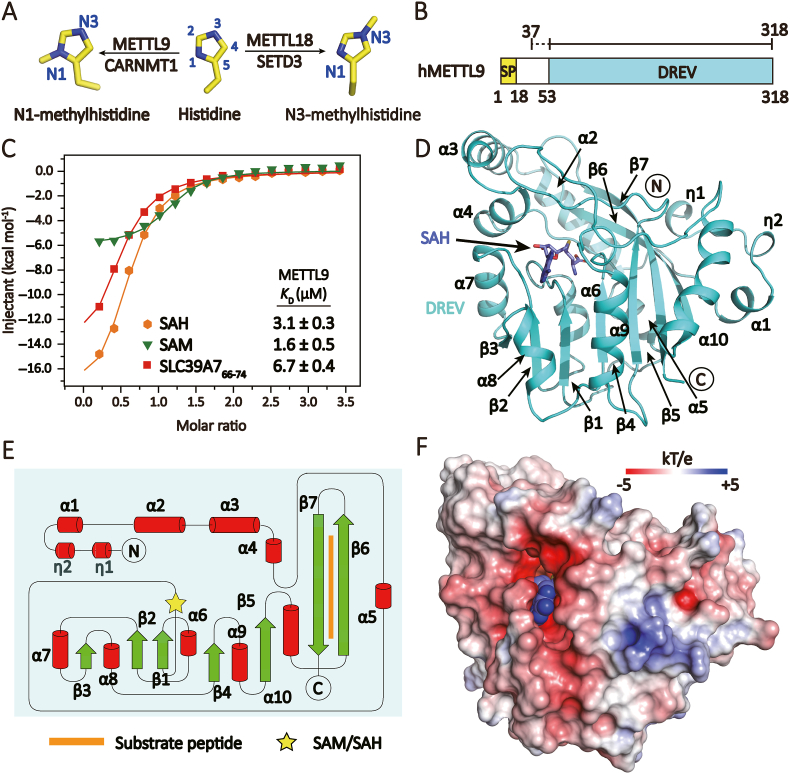
Fig. 1.
Overall structure of METTL9.(A) Position-specific histidine methylation and its writers. (B) Domain architecture of METTL9. Yellow, signal peptide (SP, residue 1–18); cyan, DREV catalytic domain (residue 53–318). The expression frame for crystallization is denoted in the top. Dashed line, invisible region. (C) Isothermal titration calorimetry (ITC) fitting curves of full-length METTL9 titrated with SAM, SAH and unmethylated substrate peptide. (D) Structure of METTL9 (cyan ribbon) bound to SAH (blue sticks). (E) Topology diagram of the METTL9 fold. Yellow star, SAH/SAM cofactors; orange line segment, substrate peptide. (F) Electrostatic surface view of METTL9 bound to SAH (blue spheres). The electrostatic potential is expressed as a spectrum ranging from −5 kT/e (red) to +5 kT/e (blue). (D) and (F) are in the same orientation. (For Interpretation of the references to colour in this figure legend the readers is referred to the web version of this article.)