-
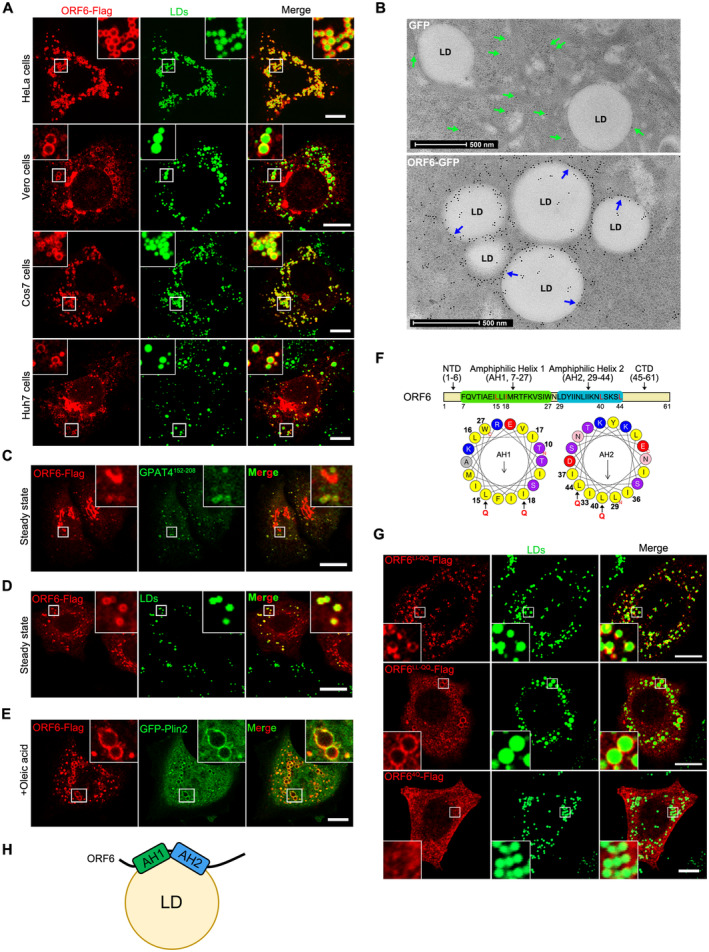
A
Cells were transfected with ORF6‐Flag for 12 h and treated with 200 μM OA for another 12 h, then stained with anti‐Flag (red). LDs were labeled with BODIPY‐493/503 (green). Cells were imaged by confocal microscopy. Scale bar represents 10 μm.
-
B
Immunogold electron micrograph of GFP or ORF6‐GFP expressed HeLa cells. Blue arrows mark ORF6‐GFP dots are enriched on the surface of LDs. Green arrows mark GFP dots are distributed in the cytoplasm. Scale bar represents 500 nm.
-
C–E
HeLa cells stable expressing ORF6‐Flag were treated with or without 200 μM OA for 12 h, then fixed and stained with anti‐Flag (red). Initial LDs were labeled with GFP‐GPAT4152–208 (green), mature LDs were labeled with BODIPY‐493/503 (green) or GFP‐Plin2 (green). Cells were imaged by confocal microscopy. Scale bar represents 10 μm.
-
F
Schematic of the domain structure of ORF6 and truncation mutants used in this study. The two predicted amphipathic helical domains of residues 7–27 (helix‐1) and residues 29–44 (helix‐2) of ORF6 were generated via the HeliQuest tool. Point mutations (black arrows) were introduced to the hydrophobic interfaces of these two helices.
-
G
HeLa cells were transfected with ORF6‐Flag mutants (L15Q, I18Q in helix‐1 or/and L40Q, L44Q in helix‐2) and were treated with 200 μM OA for 12 h, then fixed and stained with anti‐Flag (red). LDs were labeled with BODIPY‐493/503 (green). Cells were imaged by confocal microscopy. Scale bar represents 10 μm.
-
H
Schematic of the possible mechanism of ORF6 targeting to LDs.