Figure EV4. MTDPS13 patient‐derived FBXL4 variants do not efficiently assemble into an SCF complex and have impaired abilities to mediate NIX and BNIP3 turnover.
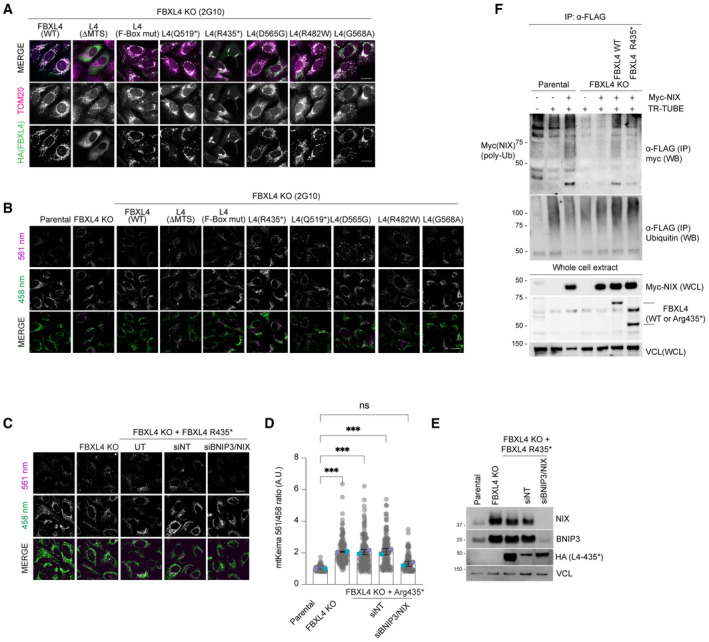
- Localisation of FBXL4 variants. FBXL4 KO cells expressing FBXL4‐HA wild‐type or specified variants were fixed and stained for HA (to detect FBXL4 in green) or TOM20 (in magenta).
- FBXL4 patient‐derived variants exhibit reduced efficiency in mediating the suppression of mitophagy compared to wild‐type FBXL4. U2OS mt‐Keima cells, U2OS mt‐Keima FBXL4 KO cells and U2OS mt‐Keima FBXL4 KO cells rescued with FBXL4 constructs were analysed by confocal microscopy. The emission signals obtained after excitation with the 458 nm laser (neutral pH) or 561 nm laser (acidic pH) are shown in green and magenta respectively. Quantification of these conditions is shown in Fig 4C.
- Elevated mitophagy in FBXL4 KO cells expressing FBXL4Arg435* truncation variant is reduced when NIX/BNIP3 are depleted using siRNA. FBXL4 KO cells stably expressing FBXL4Arg435* were transfected with siRNAs targeting both NIX and BNIP3 (NIX/BNIP3 si) or non‐targeting siRNA (NT si). Live‐cell confocal microscopy was performed to visualise mitophagy.
- Quantification of mitophagy shown in panel (C). Translucent grey dots represent measurements from individual cells. Coloured circles represent the mean ratio from independent experiments (N = 2). The centre lines and bars represent the mean of the independent replicates ± SD. At least 50 cells were analysed per condition, and over 100 cells were analysed in total per condition. P values were calculated based on the mean values using a one‐way ANOVA (*P < 0.05, **P < 0.005, ***P < 0.001, ****P < 0.0001).
- Evaluation of the extent of NIX/BNIP3 depletion in (C, D) using Western blotting.
- FBXL4Arg435* truncation variant is less efficient than wild‐type FBXL4 at promoting NIX ubiquitylation. Parental, FBXL4 KO cells, FBXL4 KO cells expressing either wild‐type FBXL4 or FBXL4‐Arg435* were transfected with TR‐TUBE and myc‐NIX, as indicated. Cell lysates obtained 48 h post‐transfection were immunoprecipitated with anti‐FLAG antibody, and the immunoprecipitates were analysed by immunoblotting using anti‐myc antibody (to detect ubiquitylated NIX or BNIP3). The line on the left marks a ladder of bands corresponding to polyubiquitylated myc‐BNIP3 or myc‐NIX.
Data information: Scale bars = 20 μm.
Source data are available online for this figure.
