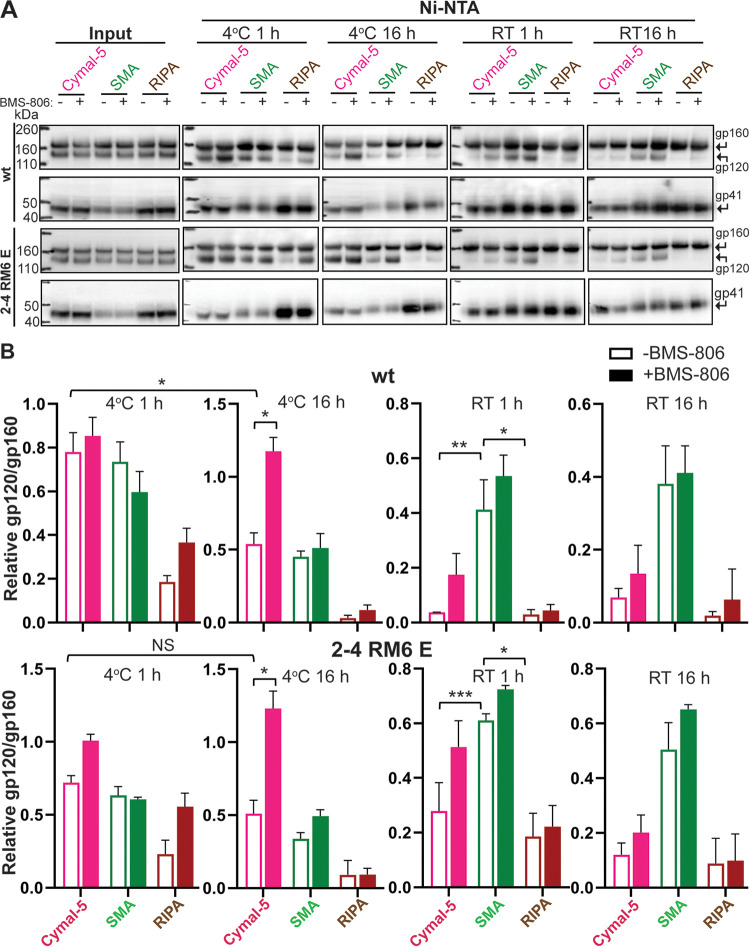
FIG 6.
Stability of Env solubilized in Cymal-5, SMA, or RIPA at different temperatures. To evaluate the integrity of the cleaved Env trimer under different solubilization conditions, we compared the relative efficiencies of gp120 and gp160 precipitation by Ni-NTA beads, which bind the His6 tags at the carboxyl termini of the Envs. (A) A549 cells were induced with doxycycline to express the wt HIV-1AD8 and 2-4 RM6 E Envs, both of which have His6 tags at the carboxyl terminus. At 48 h after induction, the cells were lysed in 1% Cymal-5, 1% SMA, or RIPA buffer in the absence or presence of 10 μM BMS-806 for 5 min on ice. After the cell debris was pelleted, a sample of the supernatant was saved as “input.” The remainder of the cleared supernatants was incubated at the indicated times and temperatures with Ni-NTA beads by end-over-end rotation. For Cymal-5, SMA, and RIPA samples, the beads were washed with 60 bed volumes of the corresponding wash buffers: (i) 20 mM Tris-HCl (pH 8.0), 100 mM (NH4)2SO4, 1 M NaCl, 30 mM imidazole, 1% Cymal-5; (ii) 20 mM Tris-HCl (pH 8.0), 100 mM (NH4)2SO4, 1 M NaCl, 30 mM imidazole, 0.005% Cymal-6; or (iii) RIPA buffer. After washing, the samples captured on the Ni-NTA beads were Western blotted with a goat anti-gp120 antiserum (upper panels) or the human 4E10 anti-gp41 antibody (lower panels). RT, room temperature. (B) Quantification of the gp120 and gp160 band intensity in A was performed in Fiji ImageJ (NIH). The stability of the Env trimer was calculated by the formula (gp120/gp160)Ni-NTA ÷ (gp120/gp160)input. The means and standard deviations derived from two independent experiments are shown. The data were compared using a Student t test. Two-tailed P values are indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001).