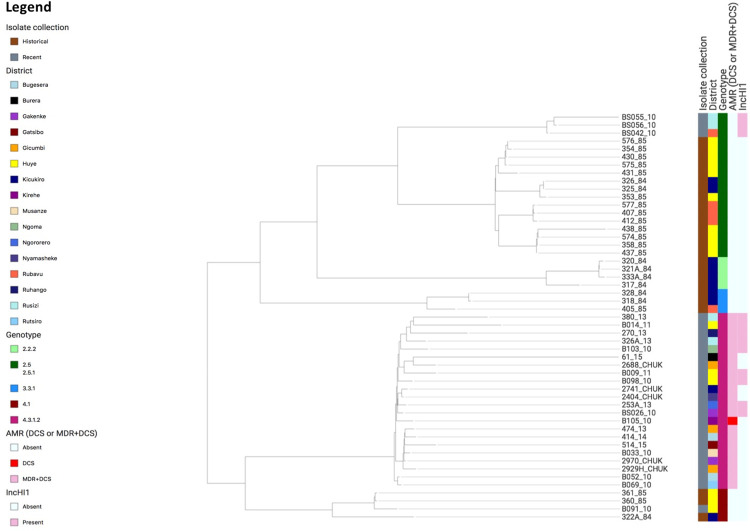
Fig 3. Phylogenetic tree of the S. Typhi isolates as generated in Pathogenwatch and visualized in Microreact (https://microreact.org/project/syLwGwmBRvbSbVeq1sET2x).
Metadata defined in the legend indicate, from left to right (i) the type of isolate collection (recent or historical), (ii) district, (iii) genotype, (iv) AMR [multidrug resistant (MDR), decreased ciprofloxacin susceptibility (DCS), or MDR + DCS), (v) presence or absence of the IncHI1 plasmid replicon per isolate.