Figure.
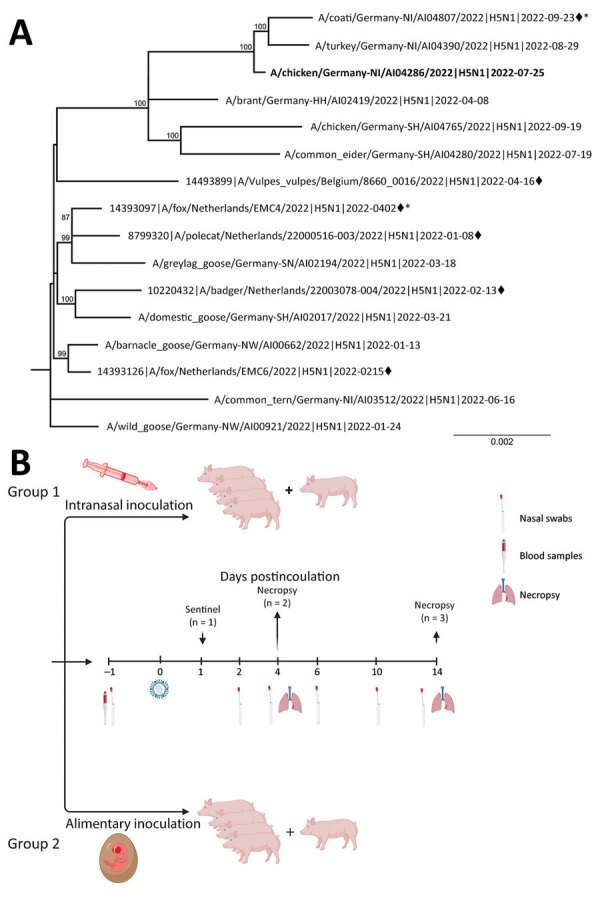
Phylogeny and experimental design for study of susceptibility of pigs against experimental infection with highly pathogenic avian influenza (HPAI) (H5N1) virus clade 2.3.4.4b. A) Maximum-likelihood phylogenetic tree (RAxML, https://cme.h-its.org/exelixis/web/software/raxml) based on 8 concatenated genome segments of selected recent HPAI H5N1 viruses from naturally infected avian hosts and from mammalian hosts (black diamonds) in Europe. Bold indicates study isolate A/chicken/Germany/AI04286/2022. Asterisks (*) indicate sequences with polymerase basic 2 E627K mutations. B) Scheme of the experimental setting of HPAI H5N1 virus infection of pigs. Four pigs, 4 months of age, were inoculated with 106 TCID50 in 2 mL using mucosal atomization devices. Four pigs were each fed with 1 HPAI H5N1 virus–infected embryonated chicken egg, carrying ≈108–109 TCID50/mL of allantoic fluid. Each pig was offered an egg, separately, in a trough and observed to complete consume it. Ten-day-old eggs were inoculated with 0.2 mL of clarified amnio-allantoic fluid of egg passage 1 and incubated for 3 days or until embryonic death was evident. Eggs were chilled until fed to pigs. Panel B created with BioRender.com and licensed by the company (agreement no. UC258UM8J3). TCID50, 50% tissue culture infectious dose.
