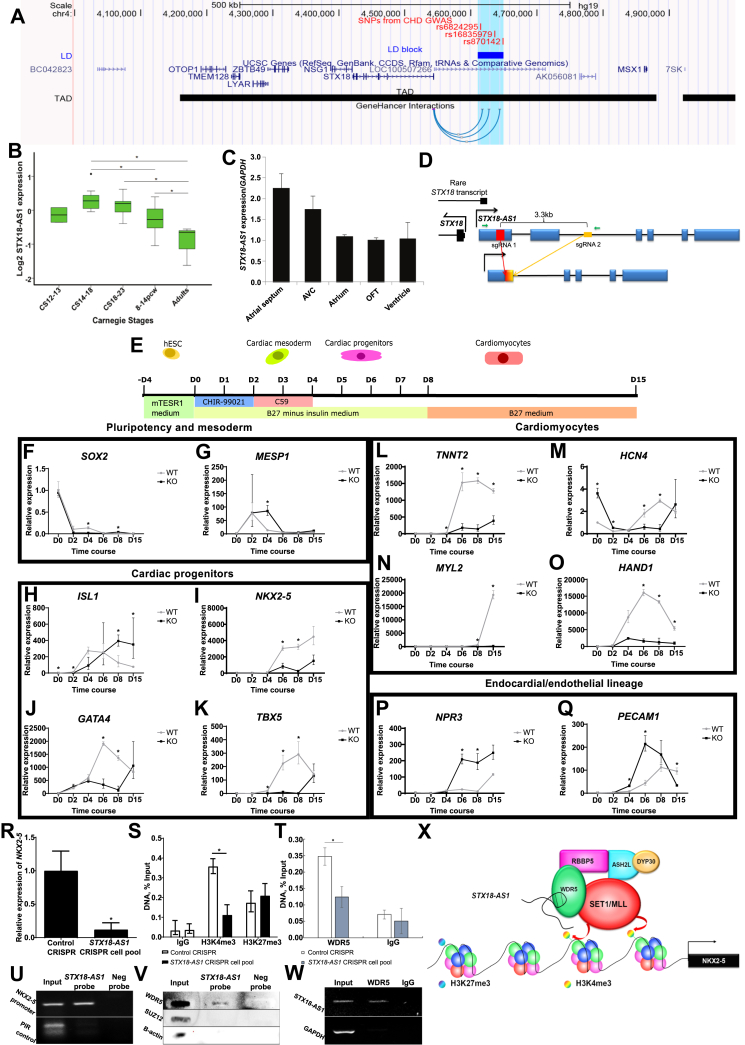
Figure 1.
Long non-coding RNA STX18-AS1 regulates in vitro cardiomyocyte differentiation via epigenetic regulation. (A) The relative genomic location of ASD-risk SNPs and STX18-AS1 (LOC100507266) with surrounding genes and chromosome interactions. GWAS identified top risk SNPs for ASD were labeled as red lines with SNP IDs. The linkage disequilibrium block (LD block in blue rectangle spares 38.8 kb) was extracted from the LD map generated with HaploView using data from 1000 Genomes Project (CEU population). Topologically associating domains (TADs) were aligned with data from 3D genome browser; while the GeneHancer Interaction (available as UCSC genome browser track) only shows the regulatory elements within the LD block region using the data from Fishilevich's paper showing the interactions between GeneHancer regulatory elements and genes. (B) Dynamic changes in STX18-AS1 transcription during human heart development. ∗, P < 0.05. 3–5 samples are included for each developmental period of Carnegie Stages (CS). (C)STX18-AS1 transcription in different human heart segments (one sample from CS15). (D) The design of the CRISPR sgRNA pair with the cuts at the first exon and second intron of STX18-AS1. The sgRNAs do not overlap any transcript of STX18. The red rectangle and yellow rectangle represent sgRNA1 and sgRNA2. The gradient colored rectangle indicates the repaired join of two cuts. (E) Time schedule and treatments applied in cardiomyocyte differentiation protocol up to Day 15, and the relevant period of cell stages from hESC to cardiomyocytes. (F, G) The time courses of markers of cell pluripotency (F, SOX2) and cardiac mesoderm (G, MESP1). (H–K) Markers for cardiac progenitors: ISL1 (H), pan-cardiac progenitor marker; NKX2-5(I), GATA4(J), and TBX5 (K), markers for cardiac progenitors and early cardiomyocytes. (L–O) Time courses of markers for cardiomyocytes: TNNT2 (L), pan-cardiomyocyte marker; HCN4 (M), specific marker of atrial cardiomyocytes; MYL2 (N), marker for ventricular cardiomyocytes; HAND1 (O), marker for cardiac mesoderm and ventricular cardiomyocytes. (P, Q) Markers of other lineages: NPR3 (P), marker for endocardial lineage; PECAM1 (Q), marker for endothelial lineage. Data are shown as Mean ± S.E. ∗, P < 0.05. Two-way ANOVA test with Bonferroni adjustment is applied for generating the P values at each time point (n = 3–9). (R)NKX2-5 transcription level was reduced in STX18-AS1 CRISPR cell pool of HepG2. (S, T) Using ChIP, H3K4me3 (S) and WDR5 (T) around the promoter of NKX2-5 was reduced in STX18-AS1 CRISPR cell pool, without changes in H3K27me3. IgG was used as a background control. (U, V) ChIRP-PCR detected the promoter region of NKX2-5 in STX18-AS1 antisense probe pulldown lysate, localizing STX18-AS1 at the NKX2-5 promoter (U). The STX18-AS1 ChIRP probes pulled down WDR5 protein but not SUZ12 and B-actin (background control) detecting with slot blotting (V). A PIR region with rare RNA binding opportunity was used as the background control. (W) RNA immunoprecipitation with anti-WDR5 pulled down STX18-AS1 RNA transcripts, detected with PCR. IgG was used as negative control antibody. (X) The model of STX18-AS1's trans-activing effects on NKX2-5 by interacting with SET1/MLL complex and regulating the histone methylation around the downstream target. Data are shown as Mean ± S.E. ∗, P < 0.05, comparing to Control CRISPR using T-test or Two-way ANOVA (n = 3).