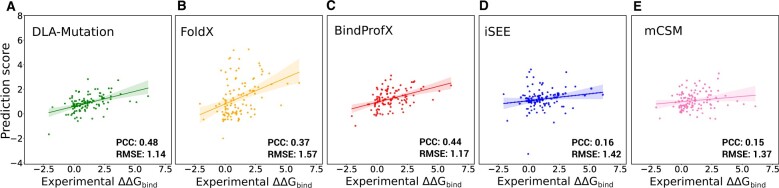
Figure 4.
Comparison between DLA-mutation and other predictors. We report values for 112 mutations coming from 17 protein complexes not seen during the training or optimization of any of the predictors. (A) DLA-mutation was trained on 945 mutations from S2003 coming from complexes sharing less than 30% sequence identity with those from this test set. We used fine-tuning of the weights and all auxiliary features. (B and E) The scores reported for FoldX (B), BindProfX (C), iSEE (D), and mCSM (E) were taken directly from Geng et al. (2019).