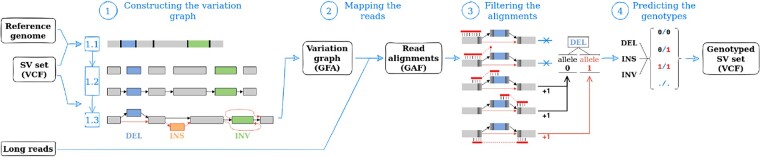
Figure 1.
Illustration of the four steps of SVJedi-graph. The method takes three files as input: the sequence of the reference genome, the VCF describing the SVs to genotype, and the long reads to genotype the SVs from. The first step is the construction of the variation graph, the second step is the mapping of the long reads on the variation graph with minigraph (producing the GAF alignment file), the third step is the filtering of the reads, and the final fourth step is the genotype prediction. Two files are output, with the main one being the genotyped version of the input VCF, and the other one being the GFA containing the variation graph.