Figure 3.
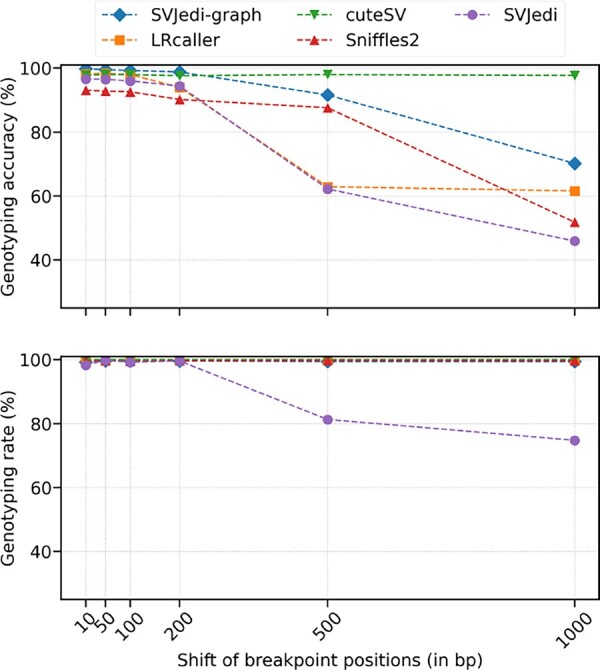
Genotyping performances of long-read SV genotypers on simulated deletion datasets on human chromosome 1 with varying levels of imprecision in the breakpoint definitions. The X axis represents the distance in base pairs between the deletions breakpoints as simulated in the input read dataset and their corresponding shifted breakpoints given in the different input VCF files.
