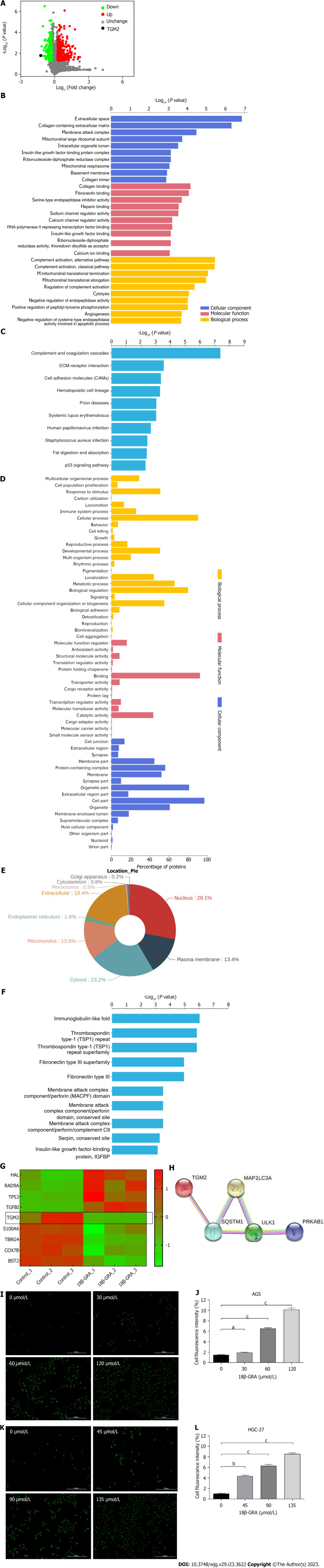
Figure 6.
The results of TMT proteomic analysis and MDC staining assay. A: TMT proteomic analysis of differentially expressed proteins (DEPs) volcano map; B: Gene Ontology (GO) enrichment analysis of DEPs. The Y axis denotes the GO functional classification enriched of DEPs, and the X axis denotes the -Log10 of the P value of Fisher’s exact test of the significance of the enrichment; C: Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The Y axis denotes the categories of KEGG pathways. The X axis is the -Log10 of the p value of Fisher’s exact test of the significance of enrichment; D: GO annotation analysis of DEPs; E: Subcellular localization analysis; F: Protein domain enrichment analysis; G: Proteins cluster analysis; H: Protein-protein interaction network diagram of TGM2 and autophagy marker proteins; I: The MDC staining results of AGS cells treated with different concentrations of 18β-glycyrrhetinic acid (18β-GRA); J: Statistical results of the MDC staining in AGS cells; K: The MDC staining results of HGC-27 cells treated with different concentrations of 18β-GRA; L: Statistical results of the MDC staining in HGC-27 cells. All data are from three independent samples. The data is represented as the mean ± SD. aP < 0.05, bP < 0.01, cP < 0.001. DEPs: Differentially expressed proteins; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; 18β-GRA: 18β-glycyrrhetinic acid.