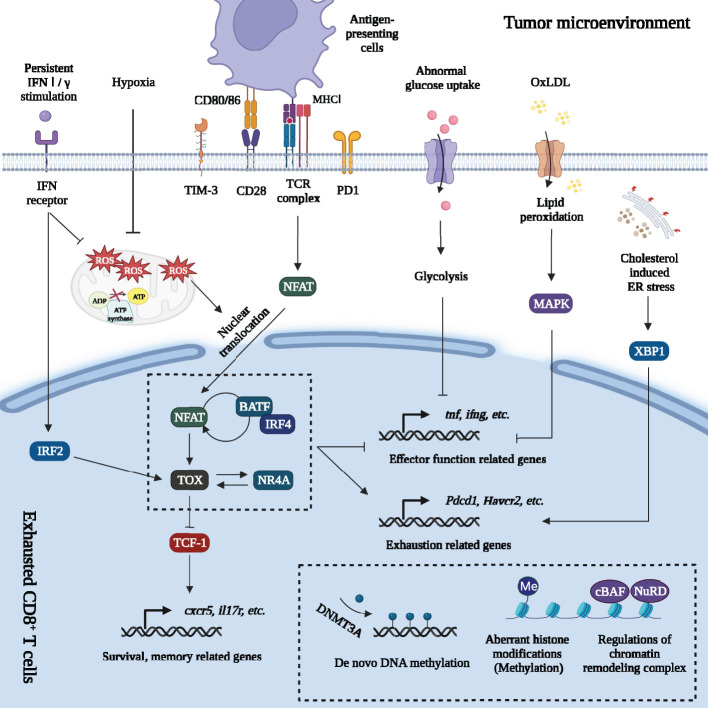
Figure 2.
Regulations of gene expression of exhausted CD8+ T cells in tumor microenvironment. Massive signals in tumor microenvironment regulate gene expression of exhausted CD8+ T cells. Firstly, persistent TCR signaling promotes NFAT dephosphorylation and nuclear translocation. The decreased of AP-1 breaks the NFAT-AP-1 cooperation but drives the NFAT-dependent program. NFAT directly enhances PD1 and TIM3 expression and promotes exhaustion. NFAT activates other TFs including TOX and NR4A. Furthermore, NFAT forms a positive feedback loop with IRF4 and BATF. TOX directly regulates or cooperates with other TFs (IRF4: BATF dimer; NR4A, etc.) to increase the expression of exhaustion-related genes and downregulate expression of genes involved in inflammatory pathways. Besides, TOX inhibits the effects of TCF-1 and promotes the terminal exhaustion differentiation. Secondly, metabolic changes also affect gene expression. Abnormal glucose uptake in TEXs suppresses glycolysis, OXPHOS and damaged effector function. Excessive intake of OxLDL leads to lipid peroxidation, which activates MAPK and inhibits Tnf and ifng expression. Cholesterol increases ER stress and upregulates the expression of inhibitory receptors (PD1, 2B4) by activating ER stress sensor XBP1. Mitochondrial dysfunction and oxidative stress greatly promote terminal exhaustion program. The impaired OXPHOS and Blimp-1 related suppression of PGC1α-dependent mitochondrial reprogramming (under hypoxia and persistent antigen stimulation) greatly increased the level of mROS. mROS was a strong inducer of terminal exhaustion differentiation. Thirdly, epigenetic changes formed the “locked” dysfunctional state through de novo DNA methylation, aberrant histone methylation modification and regulation of chromatin remodeling complex on exhaustion-related genes. Lastly, persistent IFN signaling promoted terminal exhaustion progression by inducing aberrant lipid accumulation, elevating oxidative stress in CD8+ TILs and activating IRF2. Major Histocompatibility Complex (MHC I); Nuclear factor of activated T cells (NFAT); Activator protein-1 (AP-1); Programmed cell death protein 1 (PD1); T cell immunoglobulin domain and mucin domain-3 (TIM3); Thymocyte selection-associated high mobility group box protein (TOX); Nuclear receptor 4A (NR4A); Interferon regulatory factor 4 (IRF4); The basic leucine zipper activating transcription factor-like transcription factor (BATF); T cell factor-1 (TCF-1); Oxidative phosphorylation (OXPHOS); Oxidized low-density lipoprotein (OxLDL); p38 mitogen-activated protein kinase (MAPK); Endoplasmic reticulum (ER); X-box binding protein 1 (XBP1); Mitochondrial reactive oxygen species (mROS); Interferon (IFN); Interferon regulatory factor 2 (IRF2); DNA methyltransferases 3A (DNMT3A); canonical BRG1 or BRM-associated factor (cBAF); Nucleosome remodeling and deacetylase (NuRD).