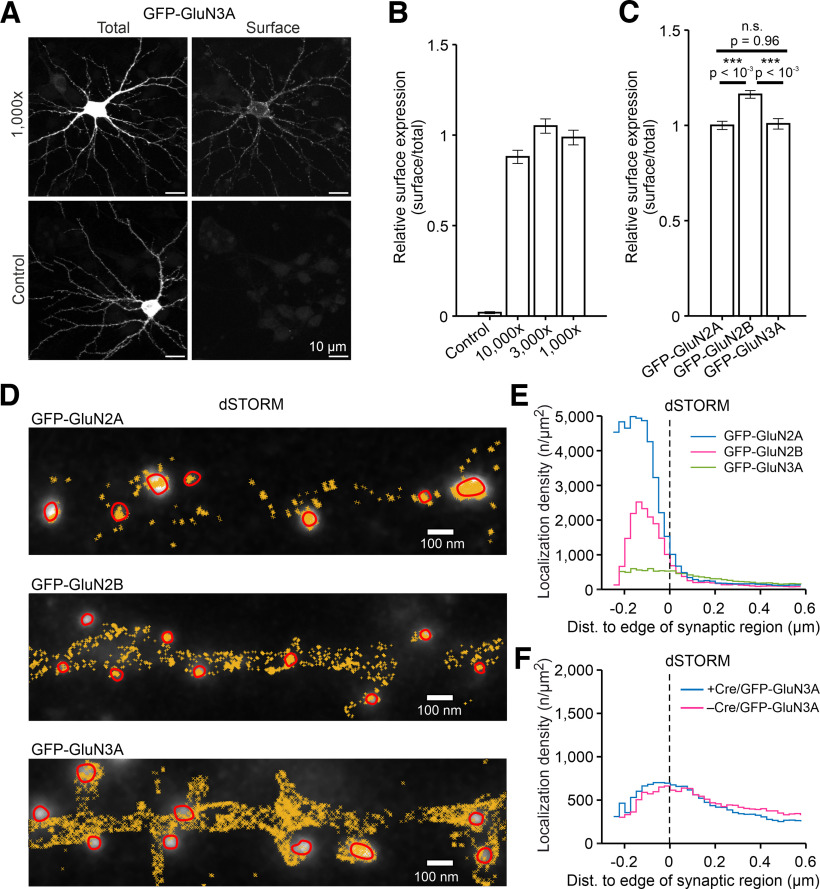
Figure 7.
Surface localization of GFP-GluN-containing NMDARs in hippocampal neurons measured by dSTORM using the nanoGFP-AF647 probe. A, Representative images of rat hippocampal neurons (DIV14) infected with GFP-GluN3A subunit; total (labeled using primary anti-GFP antibody and secondary anti-antibody conjugated with AF488) and surface (obtained using 1000-fold diluted nanoGFP-AF647 probe) signals are shown; negative control was labeled without addition of nanoGFP-AF647 probe. B, Summary of relative surface expression signals obtained by labeling with differently diluted nanoGFP-AF647 probes (1,000×; 3,000×; 10,000×) as described in A; measured in 10 μm segments of secondary or tertiary dendrites of rat hippocampal neurons infected with the GFP-GluN3A subunit (n ≥ 24 segments in ≥6 different cells/group). C, Summary of relative surface expression of indicated GFP-GluN subunits measured in 10 μm segments of secondary or tertiary dendrites of infected rat hippocampal neurons, labeled as described in A (n ≥ 24 segments in ≥6 different cells/group); one-way ANOVA (F(3,287) = 53.0405, p < 0.001 followed by Bonferroni's multiple-comparisons test with p-values denoted in the figure. D, Selected dSTORM images of rat hippocampal neurons infected with GFP-GluN2A, GFP-GluN2B, or GFP-GluN3A subunits that were labeled with nanoGFP-AF647 probe. E, Histogram of the localization density of NMDARs containing the GFP-GluN2A, GFP-GluN2B, and GFP-GluN3A subunits labeled with the nanoGFP-AF647 probe. The AF647 localizations are plotted against distance to the edge of the synaptic region, measured in rat hippocampal neurons coinfected with tdTomato-Homer1c (n > 161,793 nanoGFP-AF647 localizations/group). F, Histogram of the localization density of AF647 localizations plotted against distance to the edge of the synaptic region, measured in hippocampal neurons from cKO-GluN2A/GluN2B mice coinfected with GFP-GluN3A subunit and tdTomato-Homer1c (–Cre/GFP-GluN3A) or Cre-tdTomato-Homer1c (+Cre/GFP-GluN3A; n > 12,998 nanoGFP-AF647 localizations/group).