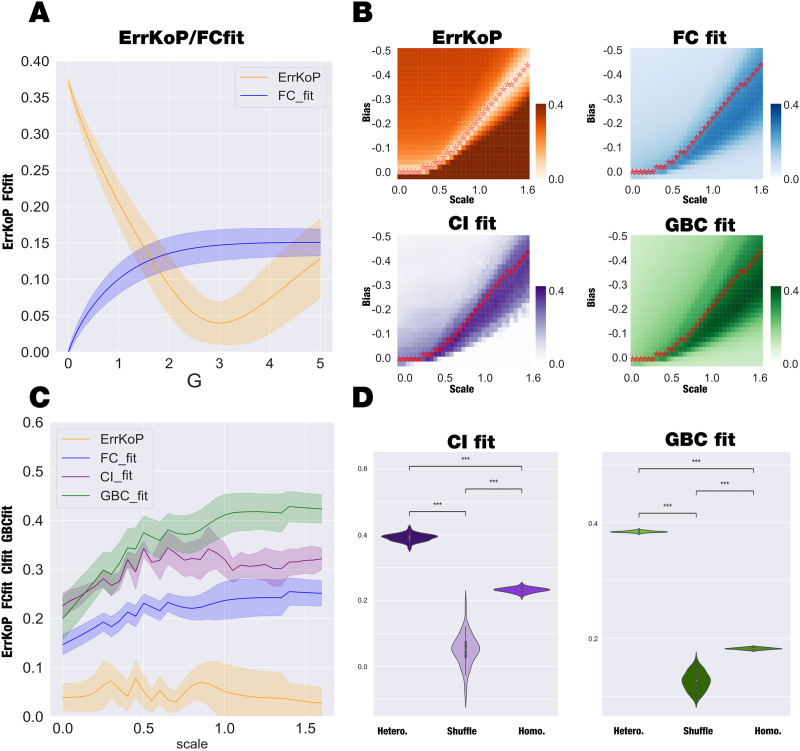
Figure 2. .
Hopf whole-brain model and the impact of regional heterogeneity T1w/T2w ratio. (A) The homogeneous model (a = −0.02 for all nodes) was fitted to the empirical data by computing the correlation between model and empirical functional connectivity (FCfit, blue) and absolute error of the model and empirical Kuramoto order parameter (ErrKoP, orange) as a function of the global coupling parameter, G. Only the fit to ErrKoP shows a clear optimum at G = 3.23, while the FC fitting level asymptotically reaches a maximum. (B) Using the optimal working point found for the homogeneous model, we introduced regional heterogeneity and assessed its impact by exploring the two-dimensional space determined by the bias and the scaling that directly modify the regional bifurcation parameters. We computed four different fitting measures: the ErrKoP and the FC, as in the homogeneous case, but also the global brain connectivity fit (GBC) and the error in Granger causality (CIfit), both computed as the Pearson correlation between the empirical and simulated measures. We defined an iso-level curve of ErrKoP, represented by red stars in the four matrices, computed as the value of bias that reaches the minimum value of ErrKoP for each value of scaling. (C) Across this iso-ErrKoP curve we computed the value of the four observables computed in panel B. Considering that the pair (0, 0) stands for the homogeneous case, we observed that CIfit, GBCfit, and FCfit increase with the inclusion of the heterogeneity (the ErrKoP remains constant by definition in this plot). (D) We selected the scale value where the maximal CIfit is reached and computed 50 times the CIfit and the GBCfit with the regional heterogeneity, with the homogeneous model, and with a spatial null model (generated by shuffling the regions of heterogeneity, preserving the spatial autocorrelation). The boxplots show the comparison of the three models in the two measures, presenting statistical significance for all the cases (*** P < 0.001, Wilcoxon rank sum test with Bonferroni correction).