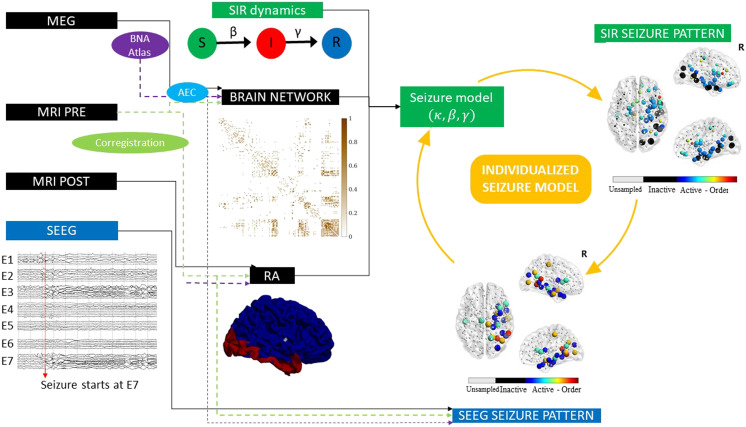
Figure 1. .
Sketch of the methodology followed in this study. The Spreading-Infected-Recovered (SIR) model was used to simulate seizure propagation. As the backbone for the model dynamics, we used the patient-specific MEG network, and the seed regions were initially defined as the resection area (RA), which was reconstructed from the pre- and postsurgery MRIs. By analyzing the seizures generated by the model, we derived the SIR seizure pattern, describing seizure propagation in the model. This was compared to the SEEG seizure pattern as derived from SEEG recordings of ictal activity. The seizure propagation patterns describe the activation order the active (i.e., infected, in the ictal state) and sampled (by the SEEG electrodes) regions of interest. Comparison between the model and the data as described in the main text allowed us to fit the model parameters to the SEEG pattern and create an individualized seizure propagation model for each patient.