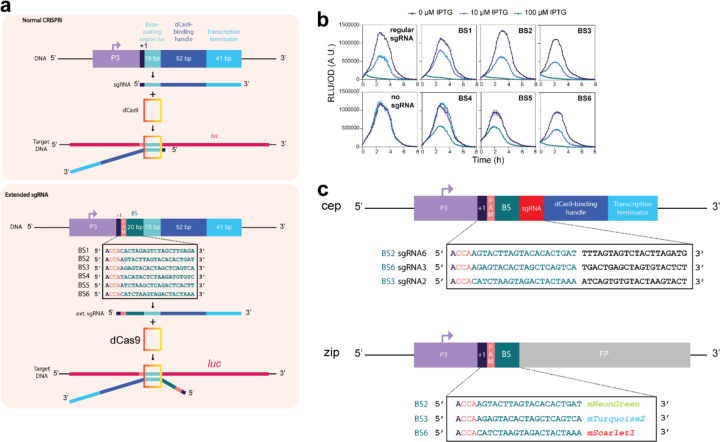
Extended Fig. 1.
Design and testing of extended sgRNAs in S. pneumoniae. (a) Normal CRISPRi uses a sgRNA that contains a 19 or 20 nt long spacer sequence that binds to a complementary DNA target sequence if this sequence also contains a PAM (top). Extended sgRNA’s (ext-sgRNAs) have a 24nt extension at their 5’ end that includes the +1, a PAM and the orthogonal binding site (BS). In the shown example, the spacer sequence is 19nt long targeting the luc gene encoding firefly luciferase (top) while the spacer sequence targeting ext-sgRNAs are 20 nts long (bottom). (b) S. pneumoniae strains harboring constitutively expressed luc and an IPTG-inducible dcas9 together with a constitutively expressed ext-sgRNA were grown in C+Y medium at 37°C in 96-well plates and OD595nm and bioluminescence was recorded every 10 min. Averages of three replicates are shown. Relative light units (RLU) over the optical density (OD) is shown on the Y-axis, time in h on the X-axis. Out of the 6 cloned and tested ext-sgRNAs, 4 showed similar luc repression levels upon dCas9 induction compared to a normal luc-targeting sgRNA. These are ext-sgRNAs containing BS1, BS2, BS3 and BS6. (c) Schematic overview of the three used ext-sgRNAs to construct the CRISPRlator and an example of how the used fluorescent reporters were constructed with a specific BS in their 5’UTR, just downstream of the +1.