Fig 1. Overall architecture of the mycobacterial Pks13 showing two alternate positions of the ACP1 domain.
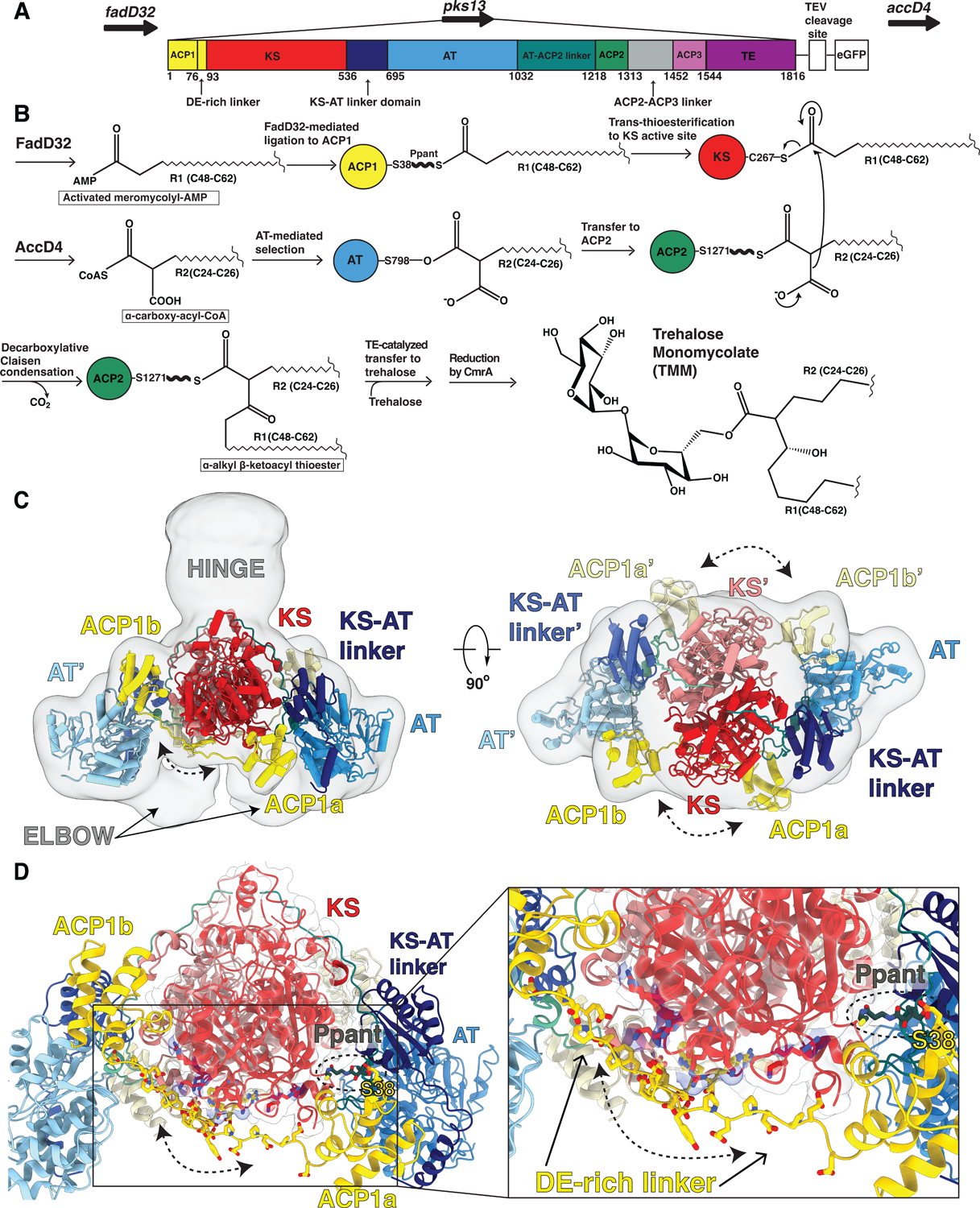
(A) Part of the Ms operon containing the fadD32 - pks13 - accD4 locus (top). Pks13 is comprised of 1816 amino acids with seven domains, and five domains of recognized function with lengths shown to scale in sequence. Colors are preserved throughout the Figures. An N-terminal acyl ACP1 (yellow) is followed by an aspartate glutamate-rich linker (DE-rich linker), KS (red), a KS-AT linker domain (navy blue), an AT (blue), AT-ACP2 linker (teal), ACP2 (green), a linker (grey), ACP3 (light purple) and TE (purple). ACP1 and ACP2 have the -DSL- motif that gets phosphopantetheinylated to receive substrate. The C-terminus of Pks13 was tagged with a TEV-cleavable eGFP. (B) Schematic of the Pks13 reaction. The meromycolyl substrate is attached to the Ppant arm (bold wave line) of the N-terminal ACP (ACP1). This is followed by trans-thioesterification to the KS. The second substrate is attached to the active site serine of AT and then transferred to ACP2 for the decarboxylative Claisen condensation. The α-alkyl β-ketoacyl thioester product is released from ACP2 by the TE that ligates it to trehalose. CmrA reduces the product to yield trehalose monomycolate (TMM). (C) A composite structure of the Pks13 homodimer showing the relative positions of ACP1a and ACP1b fitted into a composite cryo-EM map constructed from the focused reconstructions. The two protomers are distinguished from each other by darker shades of domain colors for one protomer, and lighter shades of the same color for the other. Domains in the lighter-colored protomer are labeled with the prime (‘) designation. The left ELBOW region connects the lighter colored KS and KS-AT linker domains. The right ELBOW connects the darker colored domains. The ELBOW and HINGE regions’ resolutions (~20 Å) indicate flexibility. (D) The two positions of ACP1 (ACP1a and ACP1b) are followed by acidic residues in the DE-rich linker (yellow) that lie near basic residues on the KS (highlighted blue). Ppant attached to S38 is visible in ACP1a and labeled.
