Fig 7. Proposed pathway to the structures determined for substrates bound Pks13.
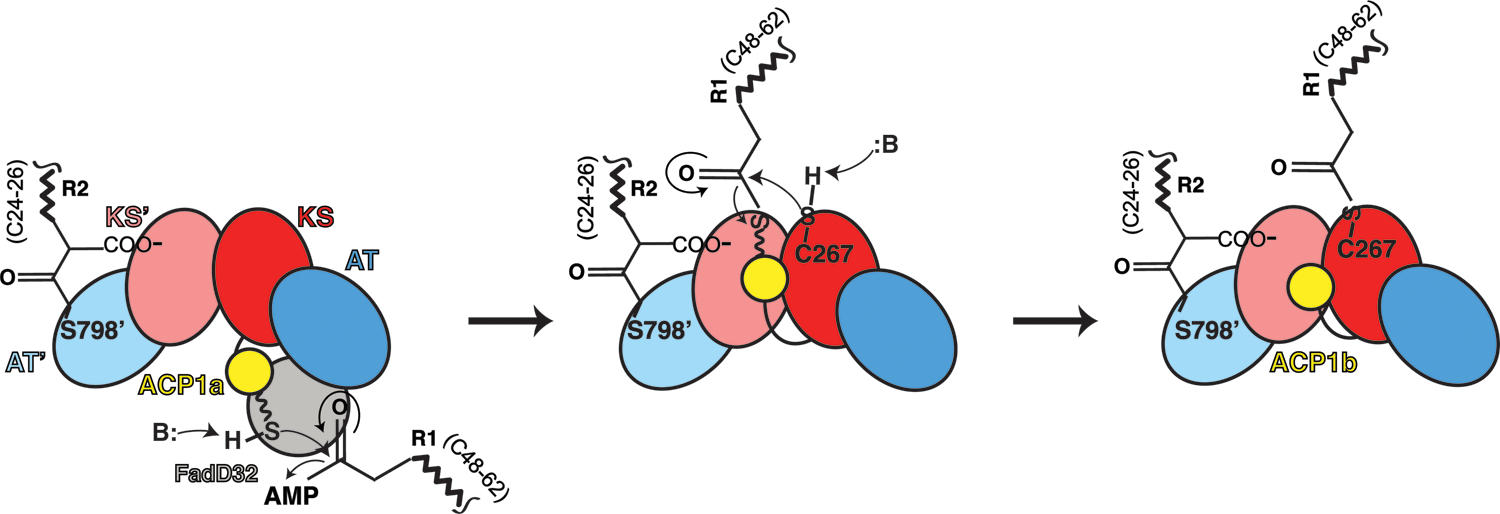
The chemical changes during the mechanism of Pks13 are schematized for just one side of the two-fold symmetric Pks13 dimer for clarity, and incorporating the two structures (ACP1a, ACP1b) we determined. The steps may occur in trans between alternate protomers as suggested by the relative proximities of the active sites of KS and AT of the contralateral protomer. Only the ACP1a, ACP1b, KS, and AT domains are cartooned to describe this initial part of the reaction up to the structure we determined (third panel). The left panel shows an early stage of the catalytic cycle in which the R2 fatty acid substrate chain of length (CH2)24–26 is attached to the AT domain as seen in the structure (Fig 5A). The Ppant arm depicted as a wavy line ending in a sulfhydryl and seen on ACP1a (yellow, first panel) links to the adenylated meromycolyl substrate (R1 (CH2)48–62) from FadD32. ACP1 delivers the attached meromycolyl substrate to the KS active site C267 through trans thioesterification as seen in the structure (third panel). This is the stable state reflected in the structure with native substrates bound. In the subsequent steps (not shown, schematized in Fig S1B) the carboxy-acyl substrate R2 is transferred to ACP2. Decarboxylative Claisen condensation proceeds between R1 and R2 where the enolate generated by decarboxylation attacks the meromycolyl thioester bound to the KS producing an α-alkyl β-ketothioester. Finally, the TE domain passes the condensed α-alkyl β-ketothioester product to trehalose and is released from Pks13. The product is subsequently reduced by CmrA to form trehalose monomycolate (TMM). TMM is subsequently transported across the plasma membrane by the transporter MmpL313.
