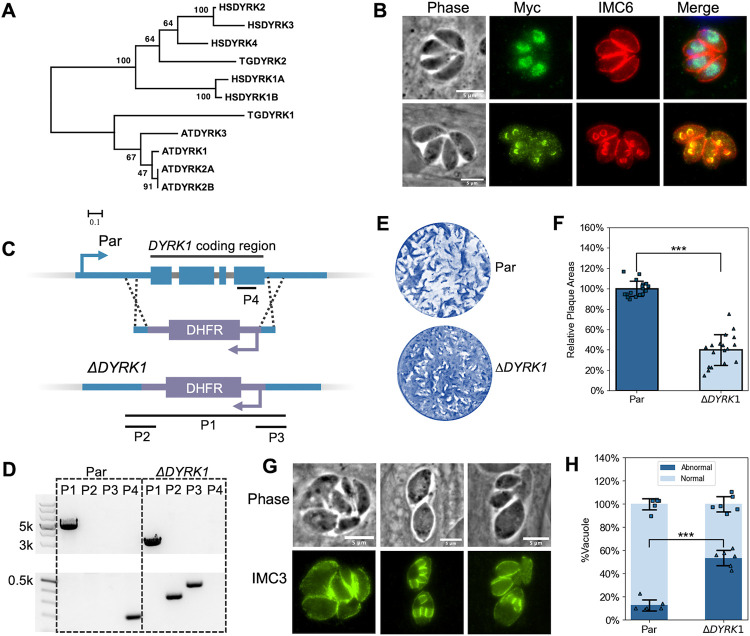
Figure 6. DYRK1 is a plant-like kinase and plays an important role in parasite division.
A. Phylogenetic analysis of DYRK sequences from humans (HSDYRK1A, 1B, 2, 3, and 4), Arabidopsis (ATDYRK1, 2A, 2B, and 3), and Toxoplasma (TGDYRK1 and 2). DYRKs from Toxoplasma are highlighted. Alignment and tree construction details and accession numbers are listed in the Methods session. B. IFA of parasites expressing Myc-tagged DYRK1 using anti-Myc and anti-IMC6 antibodies. The upper panel shows non-dividing parasites, while the lower panel shows dividing ones. Scale bar: 5 μm. C. The diagram depicts the CRISPR/Cas9-mediated strategy used to disrupt the DYRK1 gene. P1, 2, 3, and 4 are the three amplicons used to confirm the integration of the DHFR cassette between the Cas9 cutting sites and the deletion of the DYRK1 gene. D. Agarose gel of PCR products amplified from the genomic DNA extracted from ΔDYRK1 parasites using the four sets of primers indicated in C. The P1 amplicon has a length of 2928 bp in the ΔDYRK1 genome and 4787 bp in the parental genome. The P2, P3, and P4 amplicons have a length of 339 bp, 431 bp, and 196 bp, respectively. E and F. ΔDYRK1 and parental strain parasites were allowed to form plaques in culture for six days. A representative plaque assay (E) and the quantification are shown (F). The bars represent the relative average plaque areas. Three biological replicates were performed, and six experimental replicates were included each time. For each biological replicate, the data was normalized to the average plaque area of the parental. G. IFA images of ΔDYRK1 parasites that show abnormal division. H. Quantification of the ratio of vacuoles containing parasites displaying abnormal division or morphologies. *** P<0.001 (Student's t-test, two tails, unequal variance).