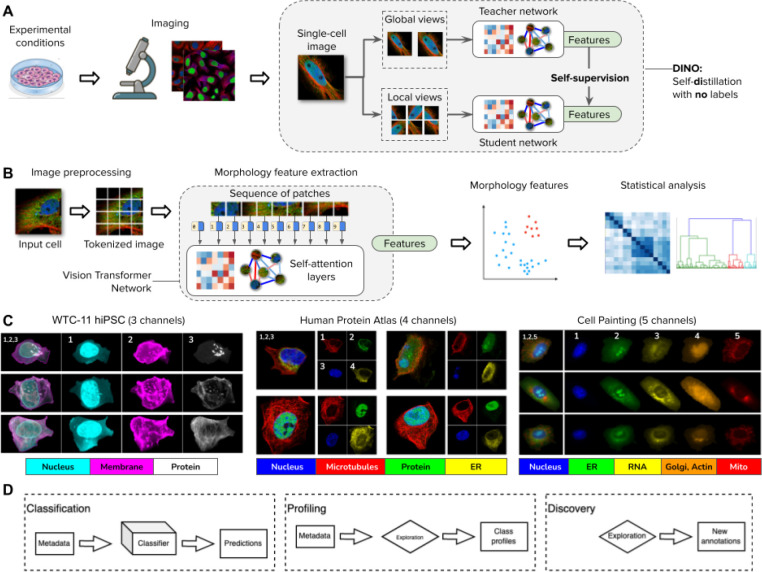
Figure 1. DINO: self-distillation with no labels, vision transformers, and datasets in this study.
A) Illustration of the workflow for collecting images of experimental conditions and then analyzing cellular phenotypes with the DINO algorithm, which trains two neural networks: a teacher and a student network. DINO is trained on images only with a self-supervised objective that aims to match the features produced by the teacher (which observes random global views of an example image) with the features produced by the student (which observes random global and local views of the same example image) (Methods). B) Illustration of the processing pipeline using vision transformer networks trained with DINO. The image of a cell is first tokenized as a sequence of local patches, which the transformer processes using self-attention layers (Methods). The vision transformer network produces single-cell morphology feature embeddings, which are used for downstream analysis using other machine learning models or statistical tests. C) Example images of the datasets used in this study: the Allen Institute WTC-11 hiPSC dataset, the Human Protein Atlas, and a collection of Cell Painting datasets. D) Biological data analysis and downstream tasks enabled by DINO features in the datasets mentioned in C.