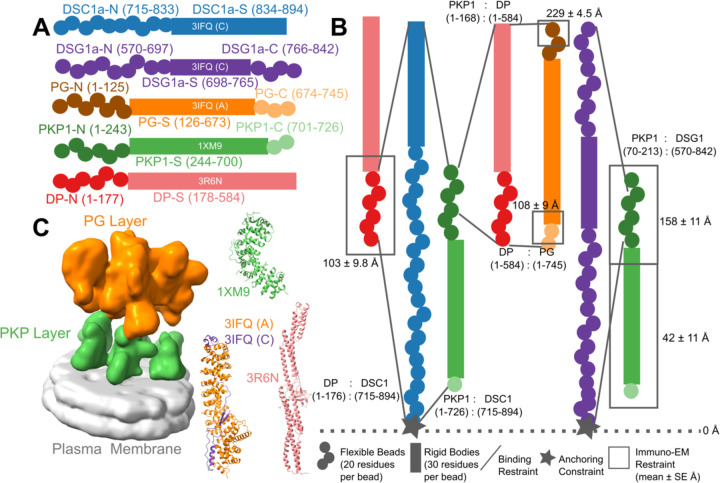
Figure 1. Representation and restraints used for integrative modeling of the desmosomal ODP.
A) The isoforms used in modeling the desmosomal ODP of stratified epithelia and the representation of the different protein domains as rigid bodies with known structure (rectangles with PDB ID and chain name) or flexible beads (circles). The domains with known structure are usually denoted by a suffix -S after the protein (e.g., DP-S), while the termini are denoted by -N or -C suffixes after the protein (e.g., DP-N). B) Three types of restraints are shown. 1. Binding restraints between interacting protein domains depicted by a pair of lines connecting the boundaries of each interacting domain pair. 2. Immuno-EM restraint for localizing protein termini depicted by rectangles around the restrained protein terminus, and 3. Anchoring constraint for localizing the transmembrane region of the cadherins depicted by star. The color scheme follows that in Panel A. C) (Left) The cryo-electron tomogram (EMD-1703) used for modeling with the PKP and the PG layers segmented. The density corresponding to the plasma membrane was not used for modeling. (Right) The PDB structures used, colored according to panel A. See also Methods, Fig. S1, Tables S1–S2.