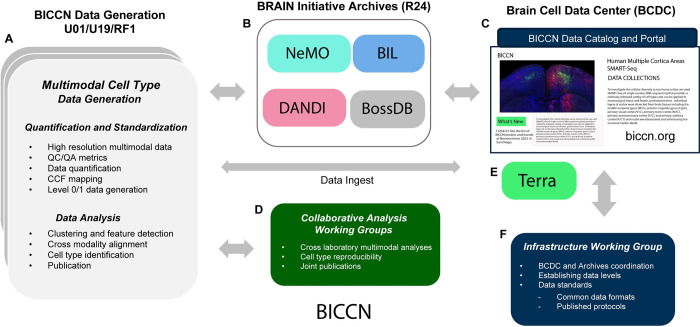
Fig 4. BICCN data ecosystem.
(A) Multimodal cell type data generation by UM1/U01/19, RF1 centers produce high-resolution Level 1 multimodal data. (B) Data are submitted to one of 4 BRAIN archives depending on data type(s): Neuroscience Multi-Omic Data Archive (NeMO), Brain Imaging Library (BIL), Distributed Archives for Neurophysiology Data Integration (DANDI) for neurophysiology data, and Brain Observatory Storage Service and Database (BossDB) for electron microscopy ultrastructural datasets. Datasets are indexed and referenced (C) by the Brain Cell Data Center (BCDC; biccn.org), which provides a portal for accessing the consortium’s data, tools, and knowledge. (D) Laboratories engage in collaborative cross-modality interpretation of data and results. (E) Terra cloud-based platform for standardized omics processing accessible through BCDC. (F) An infrastructure working group oversees architectural development and workflow management.