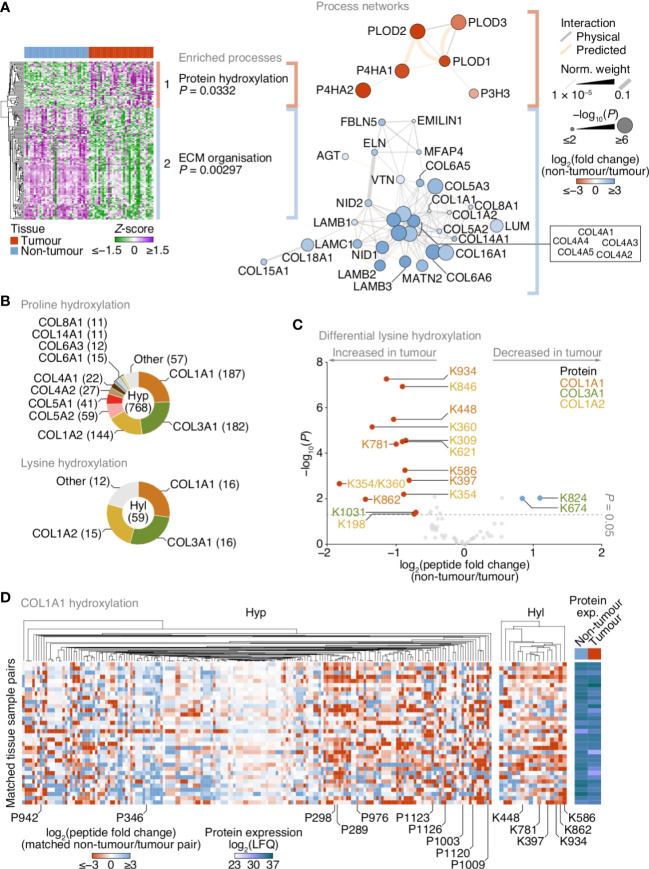
Figure 3.
Up-regulation of lysine hydroxylation in patient-derived lung tumour ECM. (A) Gene ontology enrichment analysis of the principal clusters of differentially regulated matrisome proteins identified in Figure 2 . All enriched terms for each respective cluster are shown (Fisher’s exact test with Benjamini–Hochberg correction); for cluster 2, the term “extracellular structure organisation”, the parent term of ECM organisation, and comprising the same contributing proteins and enriched with the same P-value, was omitted. Proteins belonging to enriched terms were used to construct corresponding protein interaction networks (right panels). Proteins (nodes) are coloured according to enrichment or depletion in tumour samples and sized according to statistical significance (P < 0.05, paired two-sided Student’s t-test with Benjamini–Hochberg correction). Protein interactions (edges) were weighted according to evidence of co-functionality. Unconnected proteins are not shown. (B) Proportions of identified peptides containing hydroxylated proline (Hyp; top panel) or hydroxylated lysine (Hyl; bottom panel) assigned to corresponding proteins. Numbers of modified peptides are indicated in parentheses. (C) Volcano plot of peptides containing hydroxylated lysine quantified by MS-based proteomics. Differentially regulated peptides are indicated with large coloured circles (red, increased in tumour; blue, decreased in tumour) (P < 0.05, FDR 20%, paired two-sided Student’s t-test with Benjamini–Hochberg correction). (D) Regulation of proline and lysine hydroxylation in type I collagen α1 chain (COL1A1) across 34 matched non-tumour–tumour paired tissue samples. The top ten most enriched peptides for each hydroxylation modification are labelled (all six enriched peptides containing hydroxylated lysine are labelled). Total protein expression determined by label-free quantification (LFQ) shown for corresponding samples. See Supplementary Data 6 .