Figure 1.
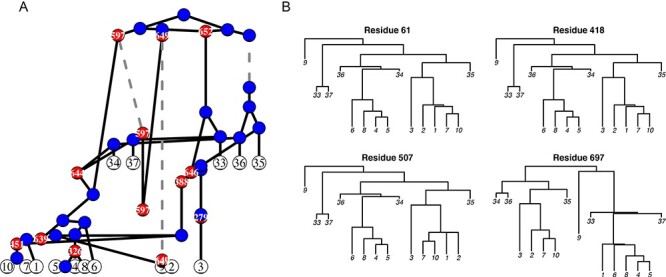
ARG decomposition into a series of bifurcating trees. (A) The simulated ARG from two sampling events. Red dots represent recombination events, with the break point identified in the white text. Blue dots represent coalescent events between two lineages. Dashed gray lines represent when a lineage is in the latent reservoir. Branch lengths correspond to time. This ARG represents the genealogical relationships among five viruses sampled 3.5 months post-infection and ten viruses sampled 7 months post-infection and was simulated using ρ = 0.08, NL = 760 assuming a sequence length of 1000. (B) Each panel shows the decomposition of the ARG into a bifurcating tree for a specific position in the alignment. Recombinant (red) nodes are resolved at the nucleotide level by removing one of the parent branches depending on where the recombination event occurred with respect to the given alignment position. If no recombination occurred between two positions, then the genealogy for those positions will be identical (e.g. Residues 61 and 418). However, recombination can alter both the topology and branch lengths of the underlying genealogy (e.g. Residues 507 and 697).
