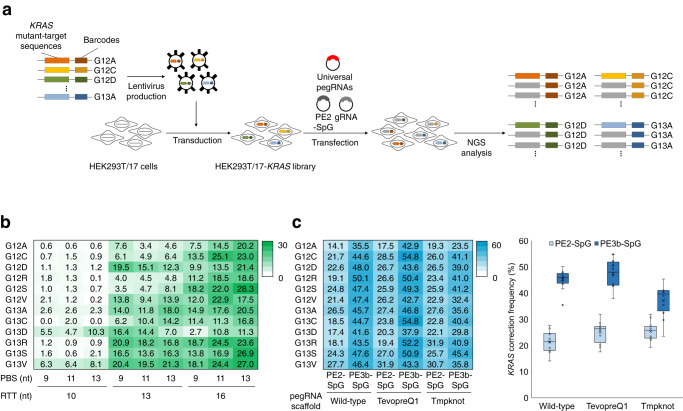
Fig. 2. Construction of HEK293T/17-KRAS library cells and correction of KRAS mutations using universal pegRNAs.
a Schematic overview for correcting KRAS mutations in HEK293T/17-KRAS library cells. Lentivirus particles containing 12 types of KRAS mutant-target sequences and barcode sequences were delivered in HEK293T/17 cells to generate HEK293T/17-KRAS library cells. After editing KRAS mutations using universal pegRNAs, the KRAS-corrected cells were collected and subjected to NGS analysis to measure the KRAS correction frequencies of each KRAS mutation. b Nine universal pegRNAs containing various lengths of PBS and RTT and PE2-SpG were delivered in HEK293T/17-KRAS library cells, and the KRAS correction frequency of each KRAS mutation is shown in the heatmap. c Universal pegRNAs containing 13-nt PBS and 16-nt RTT were engineered to have two types of RNA motif, tevopreQ1 and tmpknot, and subjected to KRAS correction in PE2-SpG or PE3b-SpG systems. The KRAS correction frequencies are shown in the heatmap and box-whisker plot, respectively. The experiments were conducted with biological triplicate (n = 3).