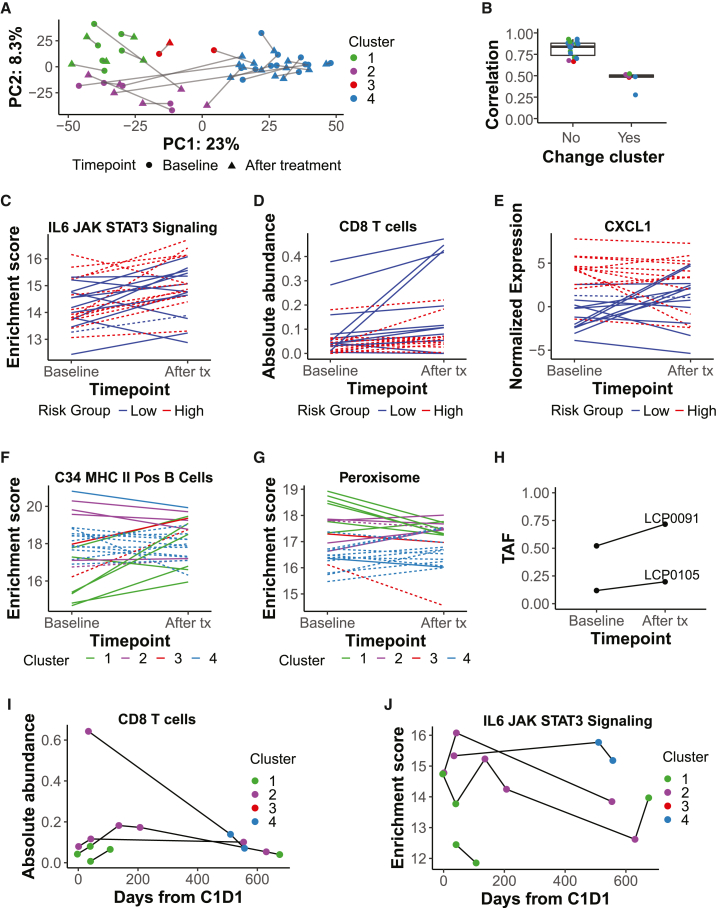
Figure 6.
Immunotherapy treatment induces an overall increase in CD8 T cells and minor differences in immune infiltration between clusters
(A) PCA on paired baseline and after-treatment tumors (n = 25 pairs) with genes used in consensus clustering as features. Baseline clusters were assigned using consensus clustering; after-treatment clusters were assigned using a k-nearest neighbors model (STAR Methods). Lines link paired tumors.
(B) Pearson correlation between baseline and after-treatment tumor pairs (on median absolute deviation [MAD] > 2 genes), split by whether the cluster changes after treatment initiation and colored by baseline cluster (see A; no, n = 19; yes, n = 6). Boxplots: center line, median; box limits, first and third quartiles; whiskers, values ≤ 1.5 times the interquartile range from box limits; points, outliers.
(C) Enrichment score over time in paired tumors for the MSigDB Hallmark IL-6 JAK STAT signaling gene set.
(D) Absolute abundance over time in paired tumors for the CIBERSORTx cell type CD8 T cell.
(E) Normalized gene expression over time in paired tumors for CXCL1.
(F and G) Enrichment score over time in paired tumors for the MSigDB c8 C34 MHC class II positive (Pos) B cell gene set (F) or for the MSigDB Hallmark peroxisome gene set (G).
(C–G) Lines are colored by the predicted risk group or molecular cluster of the baseline tumor. Dashed lines represent BTC; solid lines represent HCC. n = 25 sample pairs.
(H) Tumor allele frequency (TAF) over time for non-silent CTNNB1 mutations in two patients assigned to C1 at baseline (LCP0091, rs121913407; LCP0105, rs121913403; both known pathogenic mutations).
(I and J) For four HCC patients with longitudinal after-treatment tumor sampling, (I) absolute abundance over time of the CIBERSORTx T cell CD8 cell type and (J) enrichment score over time of the Hallmark IL-6 JAK STAT3 signaling gene set. Samples are colored by the assigned molecular cluster.