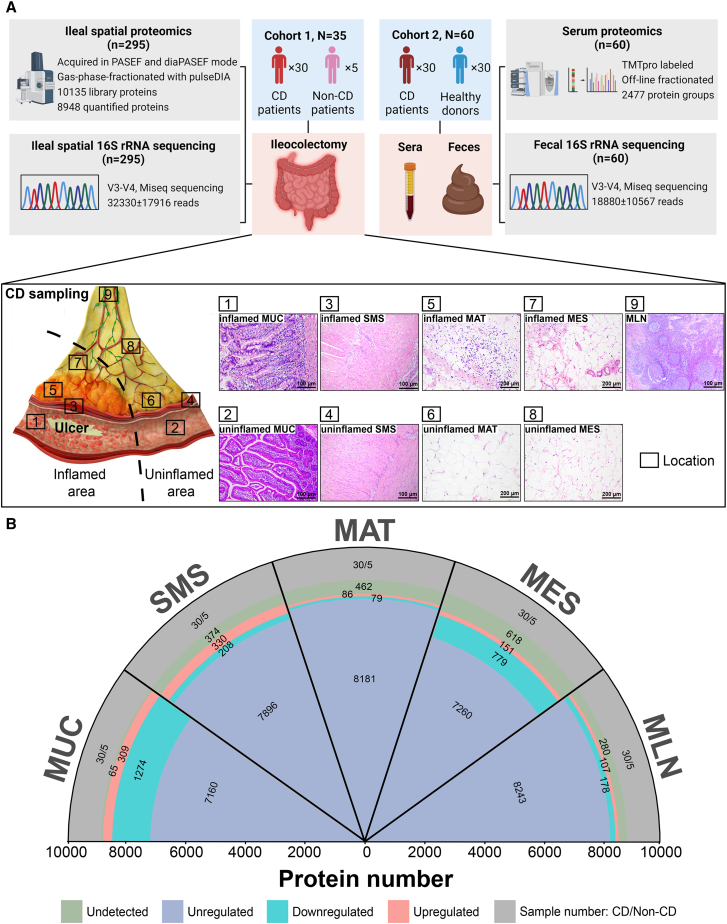
Figure 1.
The strategy for collecting multi-tissues and the general design of this study
(A) An overview of the experimental design and sample collection of the proteome and microbiome. The schematic presents the sample collection strategy of inflamed and adjacent uninflamed tissues from CD patients. iMUC, inflamed mucosa (H&E, ×100); uMUC, adjacent uninflamed MUC (H&E, ×100); iSMS, inflamed submucosa-muscularis-serosa (H&E, ×100); uSMS, adjacent uninflamed SMS (H&E, ×100); iMAT, inflamed mesenteric adipose tissue (i.e., CrF) (H&E, ×200); uMAT, adjacent uninflamed MAT (H&E, ×200); iMES, inflamed mesentery (H&E, ×200); uMES, adjacent uninflamed MES (H&E, ×200); MLN, mesenteric lymph node (H&E, ×100).
(B) Quantified and dysregulated proteins across multiple tissues. The outermost labels represent the different types of tissues (i.e., MUC, SMS, MAT, MES, and MLN). The number of samples from specific tissue is indicated in the outmost or first sector (gray). The second sector (green) refers to undetected proteins in a specific tissue. The third sector (light red) refers to upregulated proteins in a specific tissue. The fourth sector (light blue) refers to downregulated proteins in a specific tissue (B-H-adjusted p < 0.05; |log2[FC of CD versus non-CD]| > log2[1.2]), and the fifth or innermost sector (dark blue) refers to unregulated proteins in a specific tissue.