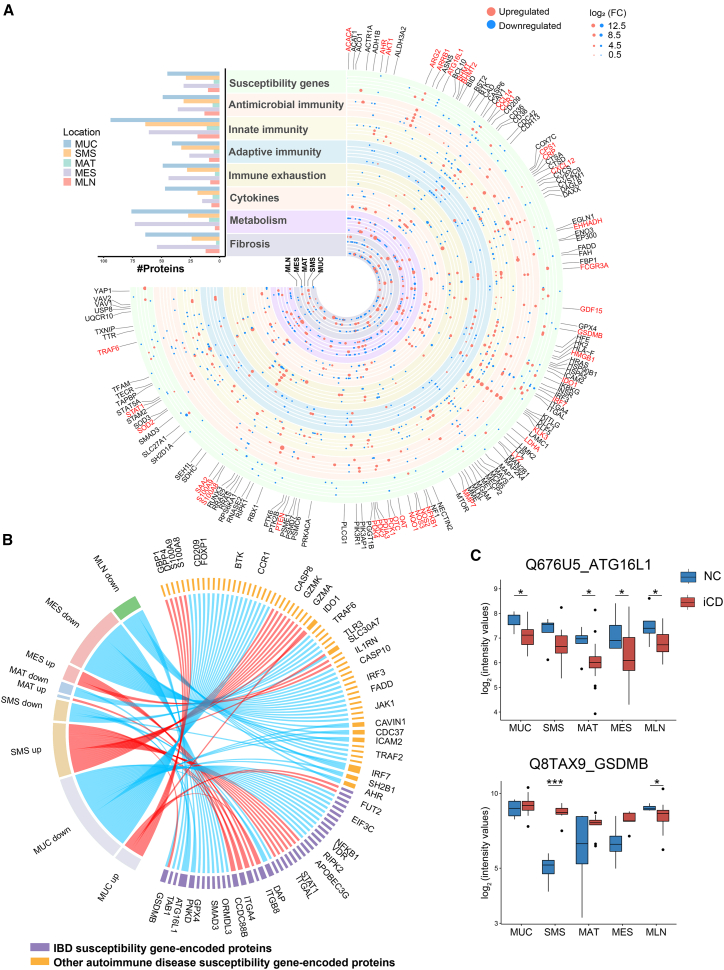
Figure 2.
Functional clusters of dysregulated proteins related to the pathogenesis of CD across multi-tissues
(A) The landscape of dysregulated proteins in multiple tissues of CD patients. Counts of dysregulated proteins in 8 clusters of protein molecules are shown in a bar chart, including IBD and other autoimmune disease-associated susceptibility gene-encoded proteins, antimicrobial immunity-associated proteins, innate immunity-associated proteins, adaptive immunity-associated proteins, immune exhaustion-associated proteins, cytokines, metabolism-associated proteins, and fibrosis-associated proteins. The horizontal columns represent the number of proteins in different functional clusters. The dysregulated proteins in 8 clusters are indicated as circles (red spots, upregulated proteins; blue spots, downregulated proteins). The size of the spots indicates absolute log2(FC).
(B) The chord diagram shows dysregulated proteins encoded by IBD and autoimmune disease-associated susceptibility genes in different tissues between CD patients and NCs. The length of the outermost brick representing each protein corresponds to the sum of |log2(FC)| in different types of tissues, and the length of the outermost brick representing each type of tissue corresponds to the sum of |log2(FC)| in one or more proteins.
(C) Protein expression of ATG16L1 and GSDMB across different tissues. The y axis indicates protein abundance. A pairwise comparison of each protein between inflamed tissues of CD patients and normal tissues of NCs was performed using a two-sided unpaired Welch’s t test. B-H-adjusted p values: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.