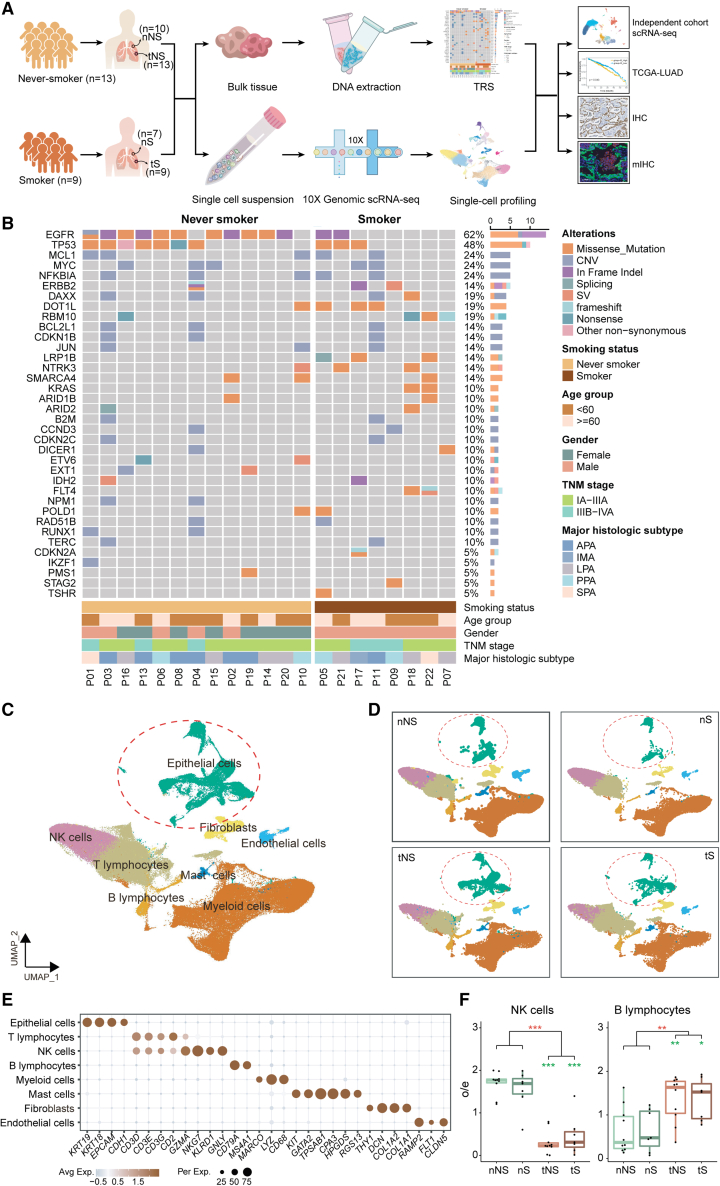
Figure 1.
The mutational landscape and comprehensive single-cell atlas of never-smoker and smoker LUADs
(A) Illustration of the workflows in this study. nNS/nS, normal lung tissue from never-smoker/smoker; tNS/tS, tumor tissue from never-smoker/smoker; TRS, targeted region sequencing; scRNA-seq, single-cell RNA sequencing; TCGA, The Cancer Genome Atlas; IHC, immunohistochemistry; mIHC, multiplex immunohistochemistry.
(B) Mutational landscape of never-smoker and smoker LUADs included for scRNA-seq analysis. The frequencies of genetic alterations are shown on the right and presented in a decreasing order. The patient IDs are presented at the bottom. The colors denote different types of genetic alterations.
(C) UMAP view of 165,753 cells from 22 patients, colored by eight major cell types; epithelial cells are circled.
(D) UMAP view of cell types in different types of tissues.
(E) Dot plot of mean expression of canonical marker genes for eight major cell types from LUADs.
(F) Tissue preference of NK cells (left) and B lymphocytes (right) in tumor tissues and paired normal lung tissues from never-smokers (n = 10) and smokers (n = 7). The observed-to-expected (o/e) ratio is the relative score of observed cell numbers over expected cell numbers calculated by chi-squared test. Each box represents the interquartile range (IQR) between the 25th and 75th percentile with the mid-point of the data, and whiskers indicate the upper and lower value within 1.5 times the IQR. Dots represent different patients. Paired two-sided Wilcoxon signed-rank test, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Asterisks represent the significance of differences in the following comparisons: all tumor tissues vs. all normal tissues (red); nNS vs. tNS and nS vs. tS (green).