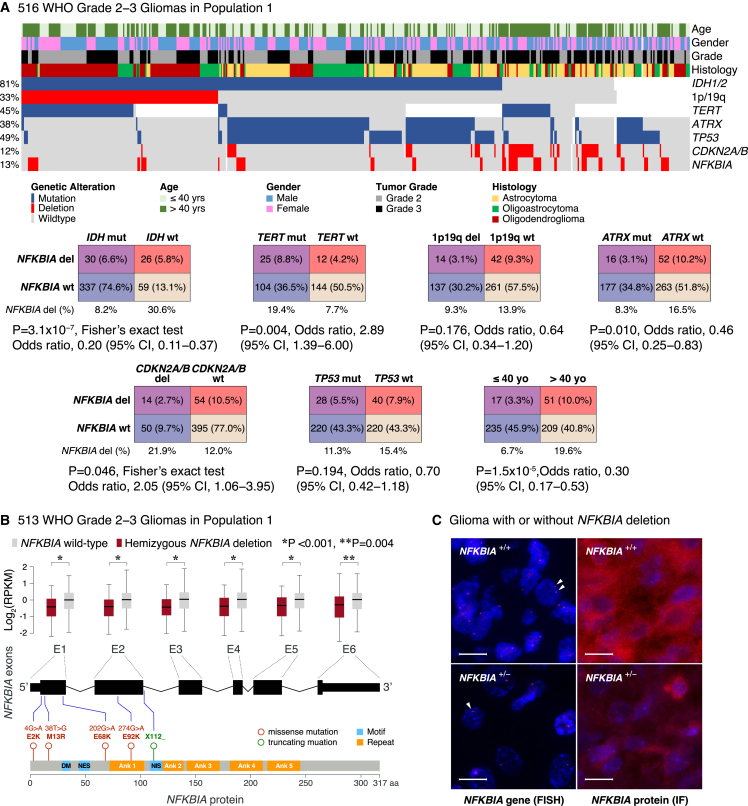
Figure 1.
NFKBIA deletions and expression in lower-grade gliomas
(A) Heatmap and two-way contingency tables showing the relationship between NFKBIA deletions and genetic alterations—IDH, TERT, ATRX, and TP53 mutations, 1p19q codeletions, and CDKN2A/B deletions—and clinicopathologic variables in 516 lower-grade gliomas of population 1. OR, odds ratio; CI, confidence interval.
(B) Expression of the six NFKBIA exons—in log2-transformed RPKM (reads per kilobase of exon model per million mapped reads)—in 513 NFKBIA deleted vs. wild-type lower-grade gliomas of population 1. Five rare missense or truncating mutations are mapped in relation to the exons and various NFKBIA protein motifs (DM, destruction motif; NES, nuclear export signal; NIS, nuclear import signal) and ankyrin (Ank) repeats. Error bars represent ±standard deviation. Wilcoxon rank-sum test.
(C) NFKBIA protein expression—by immunofluorescence (IF)—in two glioma samples with (+/–) and without (+/+) hemizygous NFKBIA deletion—by fluorescence in situ hybridization (FISH), indicating a haploinsufficiency effect. Red dots (arrowheads) represent one NFKBIA allele. Scale bars, 20 μm.