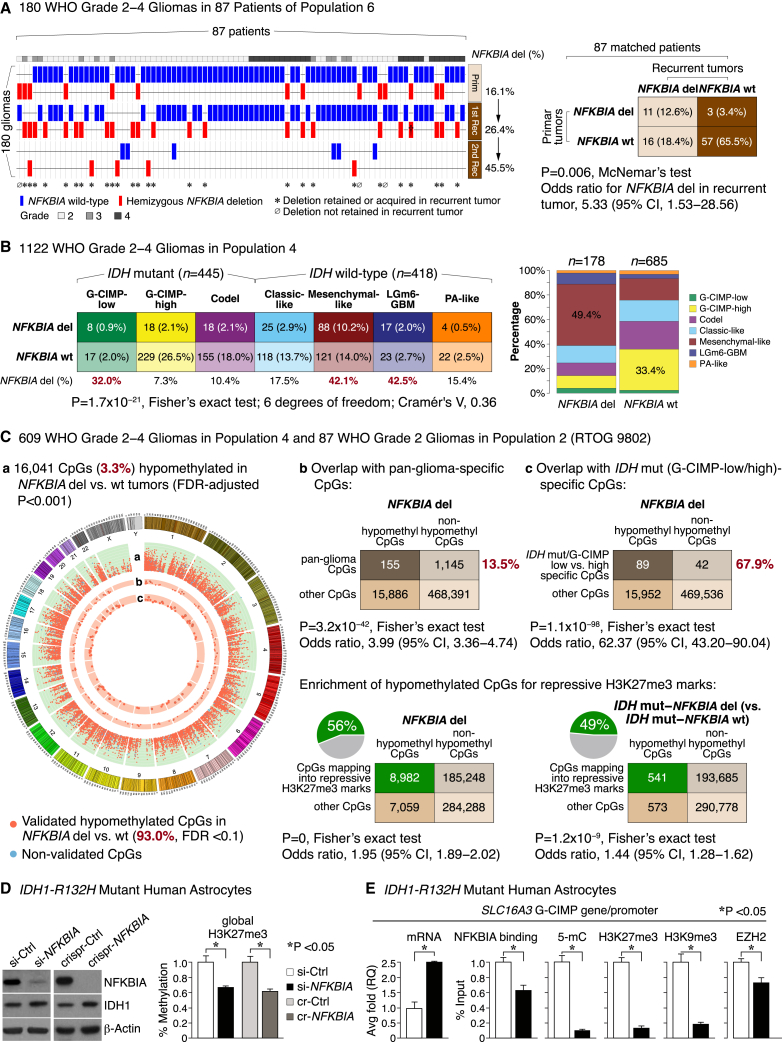
Figure 4.
The epigenome landscape of NFKBIA deleted gliomas
(A) Deletion (del) frequencies of NFKBIA in matched primary tumors vs. first and second recurrence in 180 glioma samples belonging to 87 WHO grade 2–4 gliomas in population 6. †Tumor lost a second NFKBIA allele during progression. CI denotes confidence interval.
(B) Contingency table of relationships between NFKBIA deletions and seven methylation subtypes of glioma in 1,122 WHO grade 2–4 gliomas of population 4. Cramér’s V indicates effect size. Panel on the right shows frequencies of these methylation subtypes according to NFKBIA status.
(C) Circular genome (CIRCOS) visualization of DNA regions (or CpG sites) hypomethylated in NFKBIA deleted vs. wild-type gliomas (false discovery rate [FDR] < 0.001). Data were generated in 609 WHO grade 2–4 gliomas of population 4 and validated in 87 WHO grade 2 gliomas of population 2. Orange dots in ring “a” denote genome-wide CpGs validated with an FDR of <0.1 (93.0%), blue dots denote non-validated CpGs. Orange dots in rings “b” and “c” indicate overlap of these hypomethylated CpGs with CpG signatures found to denote glioma methylation subtypes (“pan-glioma-specific”) or to distinguish G-CIMP-low from G-CIMP-high IDH mutant tumors. The height of each ring represents the beta-value—the ratio of methylated probe intensity and the sum of methylated and unmethylated probe intensities—difference range of [0.15, 0.3] between NFKBIA wild-type and deleted tumors. Dots (CpG sites) with an absolute beta difference of ≥0.2 are encapsulated by a black line. Corresponding two-way contingency tables show cosegregation between CpGs hypomethylated in NFKBIA deleted tumors and these CpG signatures. Also shown are two-way contingency tables for the cosegregation of CpGs hypomethylated in NFKBIA deleted tumors in general or in the subpopulation of IDH mutant tumors with NFKBIA deletion and CpGs that map into repressive trimethylated histone 3 lysine 27 (H3K27me3) marks. CI denotes confidence interval.
(D) NFKBIA protein expression in primary cultures of human astrocytes carrying the IDH1-R132H mutation after siRNA-mediated near-complete (si-NFKBIA) knockdown or CRISPR-mediated (crispr [cr]-NFKBIA) knockout, compared with scrambled control (Ctrl) versions. β-Actin as a loading control. Panel on the right shows global H3K27me3 levels in si-NFKBIA knockdown and crispr-NFKBIA knockout cells compared with control cells. Wilcoxon rank-sum test. Error bars represent standard error of the mean (±SEM) from three biological replicates.
(E) Bar graphs show mRNA expression—by real-time PCR average (avg) fold relative quantification (RQ)—of the SLC16A3 glioma CpG island methylator phenotype (G-CIMP) gene, its promoter binding to NFKBIA or the PRC2 catalytic subunit EZH2, and the promoter’s 5-methylcytosine (5-mC) and repressive H3K27me3 and H3K9me3 marks—all based on chromatin immunoprecipitation (ChIP) and expressed as percent of input sample representing the amount of chromatin used in ChIP—following si-NFKBIA knockdown in IDH mutant primary human astrocytes compared with their control counterparts. Wilcoxon rank-sum test. Error bars represent ±SEM from three biological replicates.