Figure 5.
NFKBIA deletion reshapes the methylome antithetically to the IDH mutation
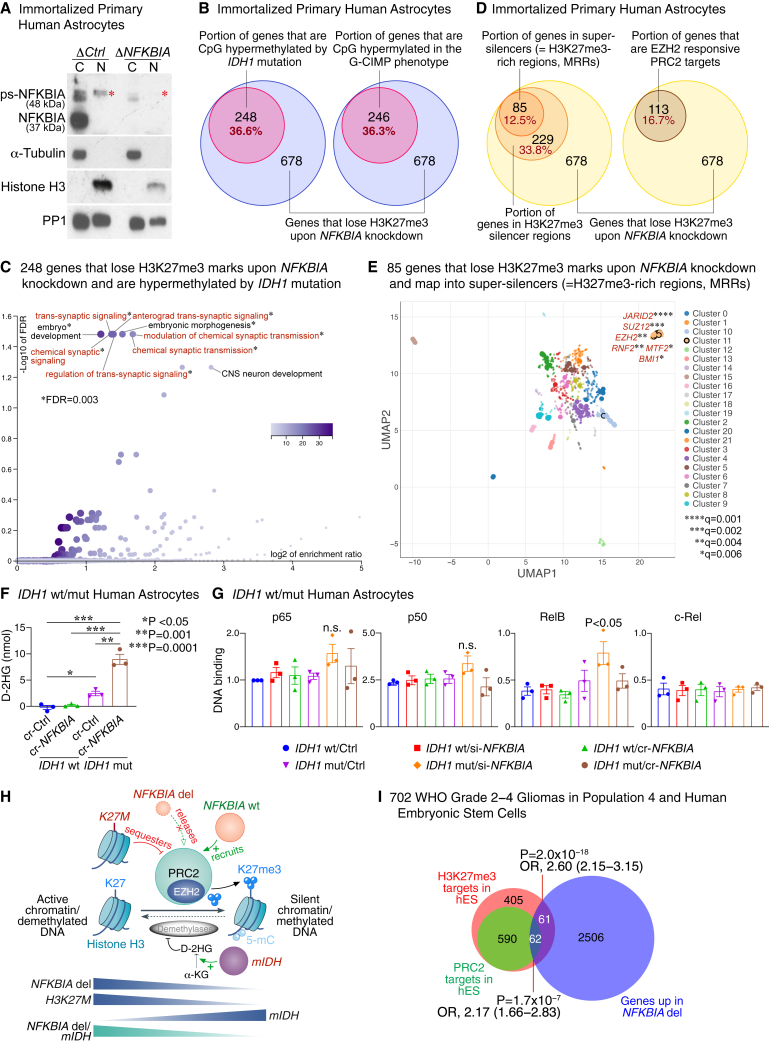
(A) Immunoblot showing expression of NFKBIA in nuclear (N) and cytoplasmic (C) fractions of control (ΔCtrl) and NFKBIA-depleted (ΔNFKBIA) immortalized primary human astrocytes. Presence of the canonical 37 kDa band in the C fraction captured with anti-phospho-NFKBIA and the ∼48 kDa band in the N fraction of ΔCtrl cells but not ΔNFKBIA cells; the latter is compatible with chromatin-bound, phosporylated-sumoylated NFKBIA (ps-NFKBIA) (red asterisks). α-Tubulin as a cytoplasmic and histone H3 as a nuclear loading control. Protein phosphatase 1 (PP1) as a loading control for both soluble and chromatin fractions.
(B) Euler diagrams showing the proportion of 678 genes with loss of repressive H3K27me3 marks by ChIP-seq upon NFKBIA depletion in primary cultures of human astrocytes and intersection with genes CpG hypermethylated upon introducing the IDH1 mutation into the same astrocyte system or with genes CpG hypermethylated in the G-CIMP phenotype.
(C) Functional enrichment analysis of the 248 genes in (A) that lose H3K27me3 marks upon NFKBIA depletion and are hypermethylated by the IDH1 mutation.
(D) Euler diagrams showing the proportions of the same 678 genes that map into H3K27me3 silencer regions or super-silencers (H3K27me3-rich regions, MRRs), and those that are EZH2-responsive PRC2 targets.
(E) Transcription factor binding site analysis for the 85 MRR-associated genes in (D) that overlap with the H3K27me3-depleted gene set. Scatterplot of terms in the ChIP enrichment analysis (ChEA) gene set library. Terms are plotted based on the first two uniform manifold approximation and projection (UMAP) dimensions. Terms with more similar gene sets are closer together. Terms are colored by automatically identified clusters computed with the Leiden algorithm applied to the term frequency-inverse document frequency (TF-IDF) values. Darker and larger points signify more substantial enrichment. Significant enrichment (points encapsulated by a black line) for components of the PRC2 core complex (SUZ12 and EZH2) and accessory proteins that define PRC2 subcomplexes PRC2.1 (MTF2) and PRC2.2 (JARID2), and other polycomb group (PcG) genes (BMI1 and RNF2). q values calculated by the Benjamini-Hochberg method.
(F) Levels of D-2-hydroxyglutarate (D-2HG) assessed by colorimetric enzymatic assay in primary cultures of IDH wild-type (wt) or IDH1-R132H mutant (mut) human astrocytes with CRISPR-mediated (cr-NFKBIA) or without (cr-Ctrl) NFKBIA depletion. Student’s t test. Error bars represent ±SEM from three biological replicates.
(G) DNA-binding activity of NF-κB proteins p65, p50, RelB, and c-Rel assessed by DNA-binding ELISAs in IDH wt vs. IDH1-R132H mut primary human astrocytes transfected with siRNA (si)-NFKBIA or cr-NFKBIA vs. Ctrl. Multiple-comparison one-way ANOVA with post hoc Tukey’s test. Error bars represent ±SEM from three biological replicates. n.s., non-significant.
(H) Graphical model of the relationship between the NFKBIA deletion and IDH and H3K27M mutations and polycomb repressive complex 2 (PRC2). Chromatin-bound NFKBIA (“NFKBIA wt”) regulates differentiation-related genes by recruiting PRC2, which promotes the trimethylation of H3K27 (H3K27me3) through its catalytic subunit EZH2, a histone methyltransferase. Genomic sequences occupied by NFKBIA overlap with those regions containing high H3K27me3 levels. Deletion of NFKBIA (NFKBIA del) results in PRC2 release and, thus, loss of H3K27me3. Similarly, H3K27M mutations in diffuse midline gliomas sequester PRC2, thereby reducing global H3K27me3 levels, especially in large unmethylated CpG islands. In turn, the IDH mutation (mIDH) hypermethylates DNA (5-mC, 5-methylcytosine) and increases global H3K27me3 levels—particularly at PRC2-targeted loci—by metabolizing α-ketoglutarate (α-KG) to D-2-hydroxyglutarate (D-2HG) and competitively inhibiting DNA and histone demethylases. Tumors with both the NFKBIA deletion and the IDH mutation demonstrate methylome changes that are more similar to those with an isolated NFKBIA deletion than to those with an isolated IDH mutation.
(I) Area-proportional Euler diagrams depicting the intersection of genes overexpressed in NFKBIA deleted gliomas (NFKBIA deletion signature) in population 4 and those identified as H3K27me3 or PRC2 targets in human embryonic stem (hES) cells. OR, odds ratio.