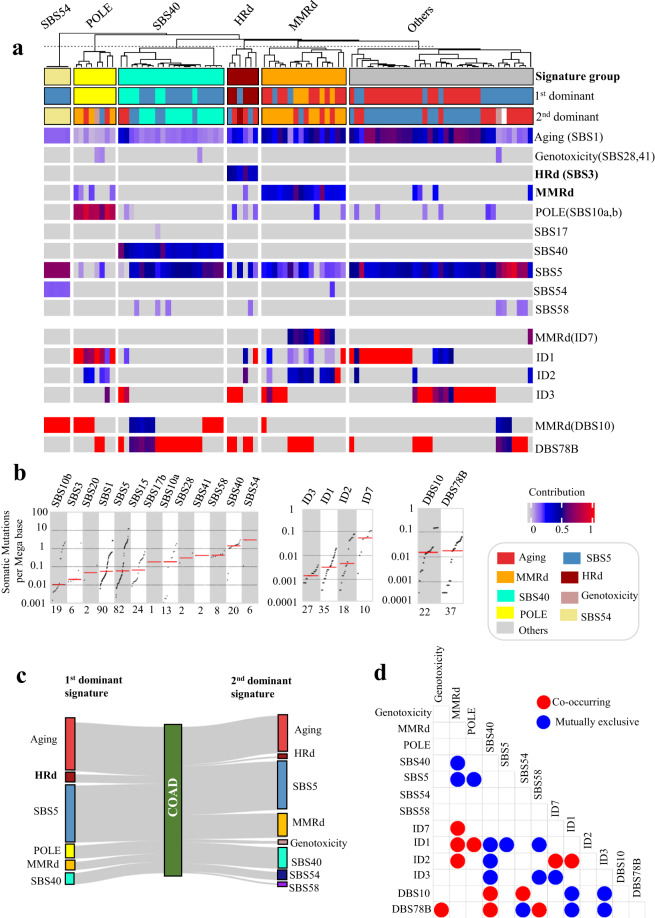
Fig. 6.
NMF-based mutational signature analysis of WGS data from COAD tumors. (a) NMF-based de novo mutational signatures of COAD WGS tumors visualized by a heatmap divided based on distinct signature status. The first and second dominant signatures are annotated at the top. Hierarchical clustering was performed based on the relative contribution of signatures in each tumor. Color codes represent each mutational signature shown. The color scale shows the contribution values of each mutational signature. For mutational signatures with known etiology, both signature and etiology are indicated. (b), (c) TMB of SBS and ID and DBS signatures for COAD tumors, respectively. TMB is measured in somatic mutations per Megabase (Mb). In the TMB plots, columns represent the detected mutational signatures and are ordered by mean somatic mutations per Mb from the lowest frequency, left, to the highest frequency, right. Numbers at the bottom of the TMB plots represent the numbers of tumors harboring each mutational signature. Only samples with counts more than zero are shown. (d) The first and second dominant SBS mutational signatures in COAD tumors. Dominant signatures were based on the contribution value of detected mutational signatures. (e) Pair-wise statistical interaction of signatures measured by hypergeometric test: red represents co-occurrence and blue represents mutual exclusivity.