Fig. 4.
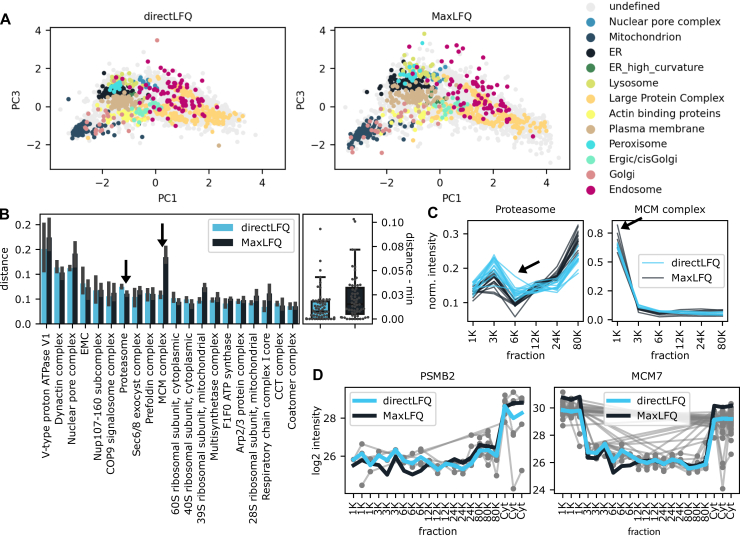
Applying directLFQ to dynamic organellar maps data from ref. (28) and comparing with MaxLFQ.A, principal component analysis maps of the dynamic organellar maps data in which protein clusters are color coded. Several clusters such as Golgi and Mitochondrion are separated more clearly with directLFQ. B, quantitative assessment of the similarity of the intensity profiles of protein clusters (lower distance means better consistency). On the left, the distances with error bars are displayed for each tested protein cluster. The arrows indicate the two clusters minichromosome maintenance (MCM) complex and Proteasome where directLFQ and MaxLFQ perform best, respectively. On the right, the normalized distances are compared with each other as boxplots; directLFQ has significantly lower distance (p = 0.014, two-sided t test). C, protein intensity profiles of these two clusters. One outlier trace in each cluster is marked by an arrow. D, visualization of the protein profiles over all replicates together with the underlying ion data. The traces show that directLFQ faithfully represents the underlying data.