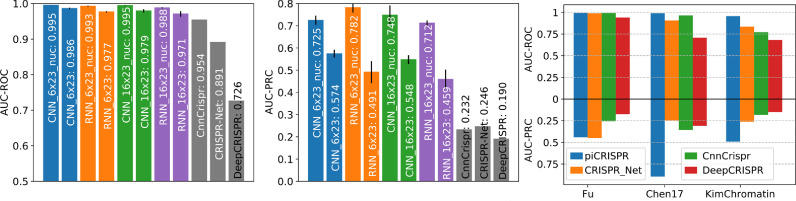
Fig. 3.
Comparison of piCRISPR models with published algorithms. All models were tested on held out studies [31], [32], [33] (testing scenario 1). Non-validated data points have been oversampled in the test set to match the class imbalance of 1:79.35 found in the dataset I-1 from [13]. piCRISPR models have been trained on the remaining data points within the crisprSQL data set. Left two panels: Comparison with three published off-target prediction algorithms [11], [13], [26] that were run on this test set. Within a model family of the same colour, the model labelled “nuc” contains nucleosomal features whereas the other does not. piCRISPR training and testing have been repeated 5 times to obtain mean and standard deviation as shown. For the underlying ROC and PRC curves see Figure S1. Right panel: AUC-ROC and AUC-PRC benchmarks for the RNN model with nucleosomal features, resolved by individual study within the held out test set.