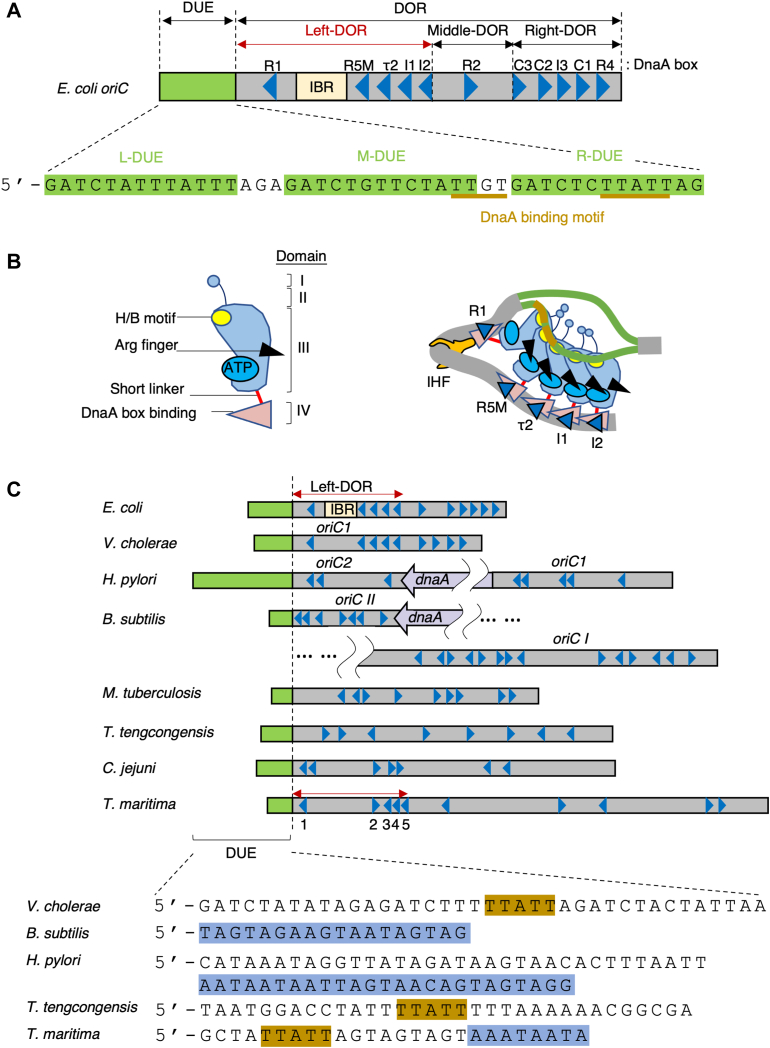
Figure 1.
Models for unwinding of the replication origin in eubacteria. A, the structure of E. coli oriC. DnaA boxes (R1, R5M, τ2, I1, I2, C3, C2, I3, C1, and R4) are indicated by arrowheads and the IHF-binding region (IBR) by a rectangle. DnaA box τ1, which overlaps IBR, is omitted for simplicity. DUE comprises L-, M-, and R-DUE (colored in green). The DnaA-binding motifs, TTGT and TTATT, are highlighted in brown bars. Left DOR spans DnaA boxes R1–I2, middle DOR consists of DnaA box R2, and right DOR spans DnaA boxes C3–R4. B, a model for open complex formation through the ssDUE recruitment mechanism. Schematic illustration of the domain architecture (I-IV) of a typical protein of the DnaA family. Domains III and IV are connected by a short linker in E. coli. The H/B and Arg-finger motifs are also indicated. In E. coli, the ATP-DnaA pentamer formed on IHF-bound left DOR unwinds DUE and concomitantly binds the upper strand of the ssDUE in a manner specific to the sequence TT[G/A]T(T). C, comparison of the overall structures of the replication origins of Eubacteria. DORs of the indicated eubacterial organisms are aligned. The bilobed structure of the origins of Bacillus subtilis and Heliobacter pylori are depicted, with one lobe, oriC II for B. subtilis and oriC2 for H. pylori, bearing the DUE. The overall structures of the origins of Mycobacterium tuberculosis (29), Thermoanaerobacter tengcongensis (30), and Camplyobacter jejuni (31) are also shown. DUE is indicated by green bars, and the sequences of experimentally characterized DUEs from B. subtilis H. pylori, T. tengcongensis, and Thermotoga maritima are shown below, highlighting the TTATT motif in brown and DnaA-trios in blue. DnaA boxes and IBR are shown as in panel A. E. coli Left-DOR and minimal tmaDOR are indicated by red left-right arrows. DOR, DnaA oligomerization region; DUE, duplex unwinding element; IHF, integration host factor; ss, single-stranded; tma, Thermotoga maritima.