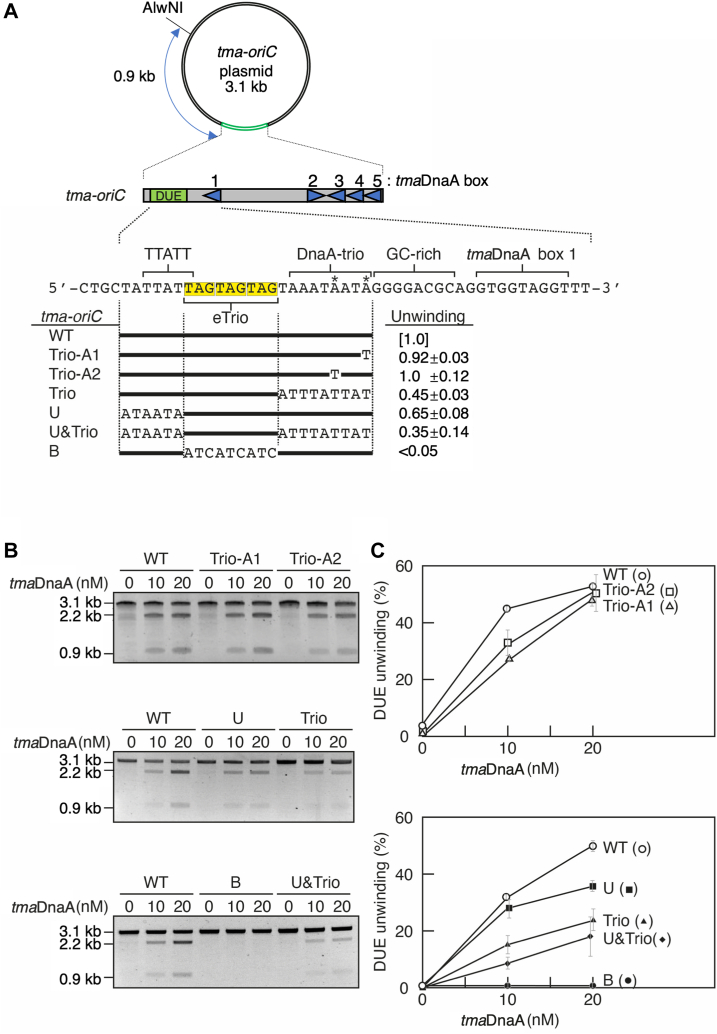
Figure 2.
The three tandem TAG repeats are essential for DUE unwinding. A, the structure of tma-oriC plasmids. pOZ14 is a pBluescript derivative with a 149 bp minimal tma-oriC DNA (WT). tma-oriC is indicated by a gray bar, tmaDUE by an green box (DUE), and tmaDnaA boxes 1 to 5 by blue triangles (box 1–5). Sequences of the TTATT, eTrio, DnaA-trio, GC-rich, and tmaDnaA box 1 motifs are shown below the tma-oriC, as are derivatives of the tmaDUE region. For mutant tma-oriC plasmids, intact DUE regions are indicated by bold lines and base substitutions by letters. The percentages of the open complex at 20 nM tmaDnaA (unwinding) are shown relative to that of the wildtype (WT). B and C, open complex formation. WT and mutant tma-oriC plasmids were individually incubated with the indicated concentrations of ATP-tmaDnaA, followed by P1 nuclease digestion. After purification, DNA samples were further digested with AlwNI and analyzed by 1% agarose gel electrophoresis and ethidium bromide staining. (B) Gel images. (C) Mean ± standard deviation percentages of P1 nuclease-digested DNA (n = 2) quantified by FIJI software. DUE, duplex unwinding element.