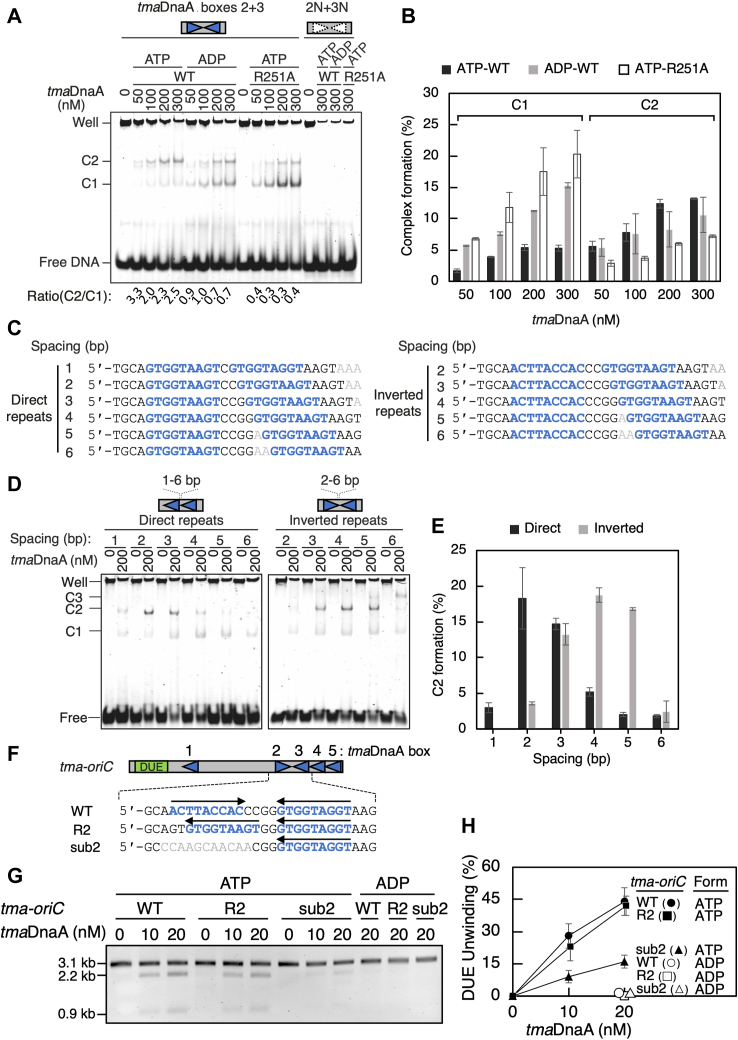
Figure 7.
Binding of tmaDnaA on DNA with two tandem tmaDnaA boxes. A and B, ATP- or ADP-bound wildtype tmaDnaA or ATP-bound tmaDnaA R251A was incubated with a 30 bp DNA (300 nM) containing tmaDnaA boxes 2 and 3 (tmaDnaA boxes 2 and 3) or with a DNA fragment lacking tmaDnaA boxes (2N and 3N). A, visualization of the tmaDnaA–DNA complexes (C1 and C2) by 8% polyacrylamide gel electrophoresis and GelStar staining. B, percentages of the tmaDnaA–DNA complexes (C1 and C2) relative to input DNA. C–E, ATP-tmaDnaA (200 nM) was incubated with a 32 bp DNA (100 nM) containing direct or inverted repeats of tmaDnaA boxes, with various spaces between boxes. C, sequences of the substrate DNA. D, visualization of the C2 complexes by 8% polyacrylamide gel electrophoresis and GelStar staining. E, percentages of C2 relative to input DNA. F–H, open complex formation. The P1 assay was performed as described in the legend to Figure 2. The tma-oriC mutant derivatives with inverted tmaDnaA box 2 (R2) or randomized tmaDnaA box 2 (Sub) are illustrated schematically (F). The gel image (G) and mean ± standard deviation percentages of P1 nuclease-digested DNA (n = 3) quantified by FIJI software (H) are shown respectively. Form, ATP- or ADP-forms of tmaDnaA. tma, Thermotoga maritima.