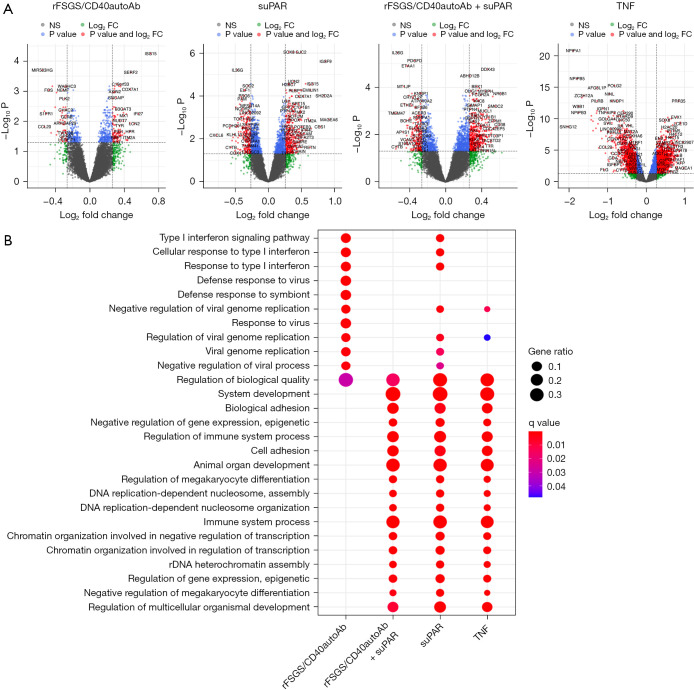
Figure 3.
Identification of molecular pathways activated in podocytes exposed to various stimuli. Microarray was performed on RNA isolated from cultured human podocytes exposed to anti-CD40 autoantibody derived from FSGS patients with recurrence of FSGS after kidney transplant (rFSGS/CD40autoAb), suPAR, a combination of suPAR and rFSGS/CD40autoAb or TNF, as described in materials and methods, was conducted. Raw data was processed using R language version 4.0.3 to obtain gene expression profiles after each treatment followed by differential gene expression and gene expression KEGG pathway analysis. (A) For each treatment volcano plot of significantly altered genes with greater than 1.2-fold change compared to untreated are depicted. (B) Results of Gene set enrichment in Gene Ontology Pathways. Differential gene analysis was performed using R package limma (2) with a P value 0.05 adjusted with Benjamini-Hochberg multiple testing correction approach. NS, genes with P value not significant. suPAR, soluble urokinase-type plasminogen activator receptor; TNF, tumor necrosis factor.