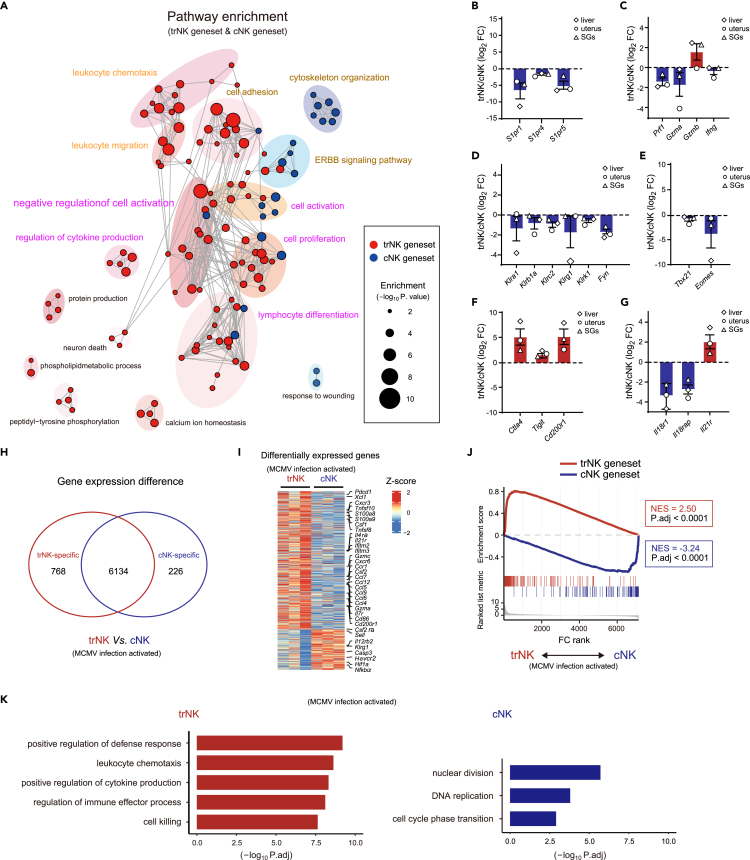
Figure 3.
trNK and cNK genesets reveal a fundamental difference between the activation of the two NK subgroups
(A) Go analysis of trNK and cNK genesets.
(B–G) Relative expression of the indicated genes between trNK and cNK in liver, uterus, and SGs, grouped by lymphocyte migration-associated molecules (B), NK effector molecules (C), NK activation-related surface receptors (D), transcription factors (E), immune checkpoints (F), and cytokine receptors (G).
(H) Venn diagram showing differentially expressed genes between liver trNK and cNK (TPM >10 in at least one group, fold change >2 and P adj. < 0.05) 44 h after MCMV infection (GSE114827).
(I) Heatmap profiling differentially expressed genes in (H).
(J) Discrimination of MCMV infection-activated trNK and cNK (H) by GSEA, using trNK and cNK genesets.
(K) GO analysis of differentially expressed genes in liver trNK and cNK after MCMV infection (H).