Figure 4. Inferred epithelial-immune crosstalk in plastic neoplastic states.
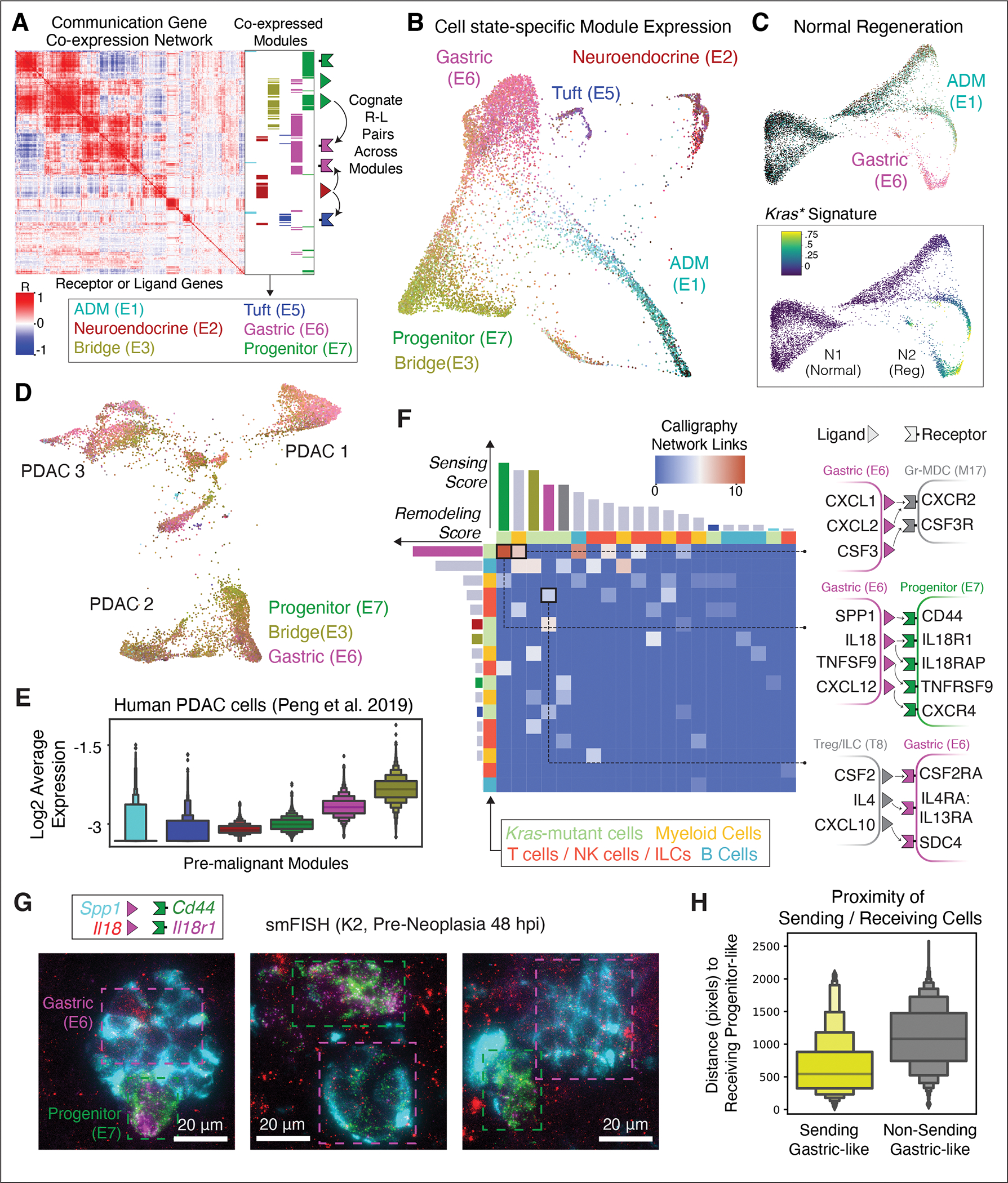
(A) Calligraphy-inferred ‘communication modules’ in pre-malignant Kras-mutant epithelial cells (K1-K3, n = 6 mice). Each row or column represents one receptor or ligand; value at intersection indicates correlation in expression (Pearson r) of that gene pair across pre-malignant cells. Blocks of highly correlated genes denote partially overlapping modules (annotated at right) that tend to co-express in the same cell-states. Schematic (far right) describes the second step of Calligraphy (see Fig. S11A). (B) FDL of K1–K3 epithelial cells with color values based on relative communication module gene expression (27). (C) FDL of KrasWT pancreas cells before and after injury (N1-N2, n = 4 mice), colored by K1–K3 communication module expression as in (B) (top) or Kras mutant signature gene expression (bottom, (23)), scaled between 1st and 99th percentile. (D) FDL of malignant cells (K5-K6, n = 3 mice), colored as in (B). (E) Communication module expression in human pancreatic tumor scRNA-seq data (32), colored as in (B). (F) Pairwise crosstalk between communication modules inferred by Calligraphy from epithelial or immune scRNA-seq data (one module per row or column), colored gray for immune or as in (A) for epithelial modules. Heat values represent number of inferred cognate R-L pair interactions across each communicating module pair; some contributing receptors or ligands are shown at right. Bars quantify total inferred edge counts, representing remodeling (row) or sensing (column) interactions for that module. (G) Two smFISH fields of view reveal the spatial proximity of sending (magenta box) to receiving (green box) mKate2+ epithelial cells. The expression of two Gastric (E6) module ligands (cyan and red), as well as two Progenitor (E7) receptors (magenta and green) overlap spatially in these three examples. Scale bars, 20 μm. (H) Distance between each receiving progenitor cell (Il18r1hi Cd44hi) and double-positive sending gastric cell (Il18hi Spp1hi), versus randomly selected non-sending gastric cells (Il18lo Spp1lo).
