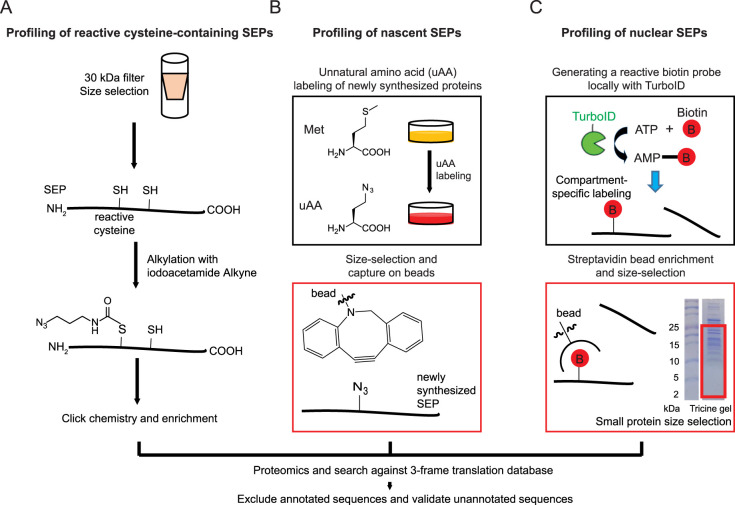
Figure 2. Chemical proteomic workflows to profile SEPs and alt-proteins.
(A) Schematic workflow of chemical proteomic profiling of reactive cysteine-containing SEPs and alt-proteins [51]. Small proteins were selected with a 30 kDa filter, followed by alkylation with iodoacetamide alkyne, which can specifically label reactive cysteine residues, to generate a handle for further Click chemistry-based enrichment. Coupled with custom smORF/alt-ORF database searching, nucleophilic cysteine-containing SEPs and alt-proteins were be identified. (B) Schematic workflow of bioorthogonal non-canonical amino acid tagging (BONCAT)-based proteomic profiling of SEPs and alt-proteins [53]. An unnatural amino acid (uAA) bearing a bioorthogonal azide moiety was metabolically incorporated into newly synthesized proteins, then the labeled small proteins were size-selected in solution with a C8 column. On-bead Click chemistry captures newly synthesized small proteins and enables removal of unlabeled proteins that did not undergo active synthesis during the labeling period, followed with trypsin digestion and proteomics to identify unannotated SEPs and alt-proteins [53]. (C) Schematic workflow of proximity labeling-based proteomic profiling of SEPs and alt-proteins [54]. TurboID, an engineered biotin ligase, biotinylates lysine residues on proximal proteins within the same subcellular region. To map SEPs and alt-proteins to subcellular compartments, TurboID was expressed in the compartment of interest as a genetic fusion to a localization sequence or protein. All proteins biotinylated by TurboID were enriched by streptavidin pulldown, followed with size-selection for small proteins and in-gel digestion (representative Coomassie-stained SDS–PAGE gel image shown). Finally, unannotated SEPs and alt-proteins were identified by mass spectrometry.