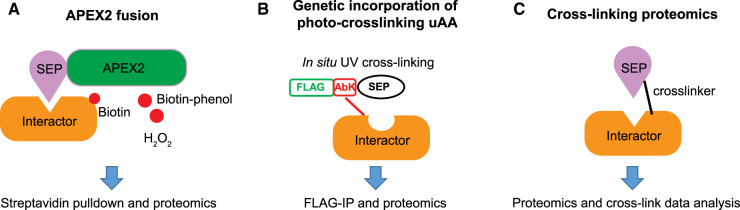
Figure 3. General workflow of chemical labeling methods to identify interaction partners of SEPs and alt-proteins.
(A) Schematic of APEX2 fusion-based proteomics to identify interaction partners of SEPs and alt-proteins. APEX2 is fused to the SEP of interest, and is expressed in cells. In the presence of biotin-phenol and H2O2, APEX2 generates biotin phenoxy radicals, which can label proximal proteins at aromatic amino acid sidechains, enabling their purification with streptavidin and identification with proteomics. (B) Schematic of photo-crosslinking uAA-based proteomics to identify interaction partners of SEPs and alt-proteins. A uAA such as AbK, a lysine analog bearing a diazirine photo-cross-linker, is incorporated into the SEP of interest using amber codon suppression technology in cells; the SEP is also fused to an epitope tag for purification of cross-linked complexes. After photo-irradiation, the diazirine is converted to a reactive carbene, which can insert into C-H bonds on proximal proteins to form a covalent bond. Immunopurification of the cross-linked complexes followed by trypsin digest and mass spectrometry enables identification of interaction partners. (C) Schematic of chemical cross-linking for identification of SEP interaction partners. Bivalent compounds containing two reactive chemical groups bridged by a linker are added to cells or lysates, enabling formation of covalent bonds to nucleophilic amino acid side chains on two interacting proteins, trapping them in a complex. Subsequent trypsin digest, followed by proteomics with smORF/alt-ORF database searching, enables global discovery of SEP and alt-protein complexes in an unbiased manner.