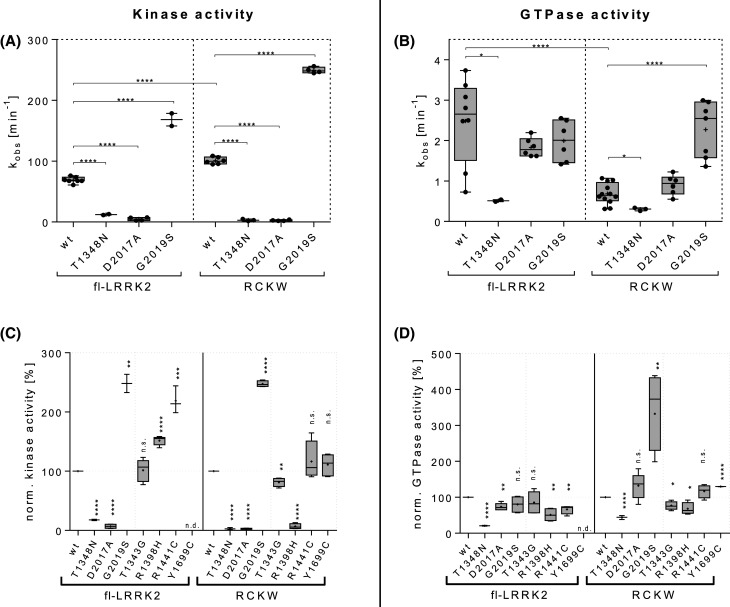
Figure 3. Biochemical and functional analysis of fl-LRRK2 and LRRK2RCKW.
(a) Absolute kinase turnover numbers kobs [min−1], determined with MMSA, show generally higher activity in LRRK2RCKW than in fl-LRRK2 regarding the wild type and the PD pathogenic G2019S mutation. Both control mutations (GTP binding deficient T1348N and kinase dead D2017A) show negligible kinase activity in both constructs. Significances were tested with parametric t-test (two-sided, confidence interval 95%) **** P < 0.0001, *** P ≤ 0.001, ** P ≤ 0.01, * P ≤ 0.05, n.s.: not significant. (b) Absolute GTPase turnover numbers kobs [min−1], determined with the radioactive charcoal assay, show generally higher activity in fl-LRRK2 than in LRRK2RCKW comparing both wild types and D2017A mutation which shows no effect on neither of the constructs. T1348N decreases GTP turnover in both constructs and G2019S increases activity in LRRK2RCKW to fl-LRRK2 levels while leaving activity unaffected in fl-LRRK2. Significances were tested with parametric t-test (two-sided, confidence interval 95%) **** P < 0.0001, *** P ≤ 0.001, ** P ≤ 0.01, * P ≤ 0.05, n.s.: not significant. (c and d) Enzymatic activities normalized to respective wild-type levels display differences in the respective effects of mutations on fl-LRRK2 and LRRK2RCKW. Most apparent are the PD related mutations R1398H, R1441C and G2019S which each increase either kinase activity (R1398H and R1441C) or GTPase activity (G2019S) in one construct while leaving the respective activity mainly unaffected (R1441C and G2019S) or decreasing it (R1398H) in the other construct (n.d.: not determined).Significances were tested with parametric t-test (two-sided, confidence interval 95%) **** P < 0.0001, *** P ≤ 0.001, ** P ≤ 0.01, * P ≤ 0.05, n.s.: not significant. A complete list of all absolute and percentage activities is shown in Supplementary Figure S1.