Michelson et al. show that the transcription factor Hnf4 upregulates gut and liver self-antigens within thymic epithelial cells via an enhancer-driven mechanism, mirroring peripheral development within the thymus to enable immunological tolerance toward the gut and liver.
Abstract
Mimetic cells are medullary thymic epithelial cells (mTECs) that mimic extra-thymic cell types to tolerize T cells to self-antigens. Here, we dissected the biology of entero-hepato mTECs, mimetic cells expressing gut- and liver-associated transcripts. Entero-hepato mTECs conserved their thymic identity yet accessed wide swaths of enterocyte chromatin and transcriptional programs via the transcription factors Hnf4α and Hnf4γ. Deletion of Hnf4α and Hnf4γ in TECs ablated entero-hepato mTECs and downregulated numerous gut- and liver-associated transcripts, with a primary contribution from Hnf4γ. Loss of Hnf4 impaired enhancer activation and CTCF redistribution in mTECs but did not impact Polycomb-mediated repression or promoter-proximal histone marks. By single-cell RNA sequencing, Hnf4 loss produced three distinct effects on mimetic cell state, fate, and accumulation. Serendipitously, a requirement for Hnf4 in microfold mTECs was discovered, which exposed a requirement for Hnf4γ in gut microfold cells and the IgA response. Study of Hnf4 in entero-hepato mTECs thus revealed mechanisms of gene control in the thymus and periphery alike.
Graphical Abstract
Introduction
Medullary thymic epithelial cells (mTECs) play a vital role in establishing T cell tolerance to self by expressing thousands of peripheral-tissue antigens (PTAs) ectopically within the thymus (Klein et al., 1998; Derbinski et al., 2001; Klein et al., 2014). Anti-PTA reactivity by a cognate T cell bearing an autoreactive T cell receptor during thymic maturation leads to clonal deletion of that specificity from the repertoire or its diversion to the regulatory T cell lineage, thus avoiding autoimmunity (Kappler et al., 1987; Kisielow et al., 1988; Bensinger et al., 2001; Jordan et al., 2001; Apostolou et al., 2002).
mTECs utilize two mechanisms to express and present PTAs to maturing T cells. The first involves the transcriptional activator Aire, which can induce the expression of myriad PTAs by redirecting general transcriptional mechanisms, including RNA polymerase II pause release, stabilization of DNA double-strand breaks, and enhancer–promoter chromatin looping (Anderson et al., 2002; Oven et al., 2007; Abramson et al., 2010; Giraud et al., 2012; Yoshida et al., 2015; Bansal et al., 2017; Guha et al., 2017; Bansal et al., 2021). Since the original discovery of Aire more than two decades ago, numerous studies have concluded that Aire-mediated PTA induction is “quasi-random,” with no apparent overarching biological logic (Derbinski et al., 2008; Villaseñor et al., 2008; Brennecke et al., 2015; Meredith et al., 2015; Dhalla et al., 2020). The second mechanism of PTA induction involves small subsets of mTECs that mimic peripheral cell types, collectively termed mimetic cells (Michelson et al., 2022b; Michelson and Mathis, 2022). As a general principle, mimetic cells co-opt diverse lineage-defining transcription factors (TFs) to drive the expression of modular sets of PTAs corresponding to diverse peripheral cell types (Bornstein et al., 2018; Miller et al., 2018; Michelson et al., 2022b; Givony et al., 2022 Preprint; Tao et al., 2023 Preprint). Aire and mimetic cells are likely closely interrelated as Aire-driven PTA expression shows some organizing principles and mimetic cell subsets depend to varying degrees on Aire for their differentiation (Brennecke et al., 2015; Meredith et al., 2015; Dhalla et al., 2020; Michelson et al., 2022b). Nonetheless, these two mechanisms operate in distinct cellular compartments and rely on distinct molecular regulators.
Aire-expressing mTECs and mimetic cells have both been shown to be necessary and sufficient sources of PTAs for T cell tolerance (Anderson et al., 2002; Devoss et al., 2006; Gavanescu et al., 2007; Taniguchi et al., 2012; Miller et al., 2018; Michelson et al., 2022b; Givony et al., 2022 Preprint). Compared with Aire-expressing mTECs, however, limited data exist on the mechanisms of PTA expression in mimetic cells. In particular, which lineage-defining TFs are required for mimetic-cell PTA expression, what molecular mechanisms these TFs use to induce PTAs, and how these mechanisms compare with PTA induction by Aire all remain largely unknown. Mimetic cells also take on many features—genomic, transcriptomic, and phenotypic—of their peripheral counterparts; nonetheless, mimetic cells derive from mTEC progenitors and do not give rise to ectopic growths or teratomas, raising questions about how lineage-defining TFs are restrained in mTECs compared with bona fide peripheral cell types (Dooley et al., 2005; Miller et al., 2018; Wells et al., 2020; Michelson et al., 2022b; Givony et al., 2022 Preprint).
To address these unknowns, we undertook a detailed characterization of one mimetic cell type, the enterocyte-hepatocyte (entero-hepato) mTEC. These mimetic cells resembled gut and liver epithelium and were critically controlled by the hepatocyte nuclear factor 4 (Hnf4) TF family. Genomic and transcriptomic studies revealed mechanisms of their differentiation and function. Serendipitously, insights derived from mimetic cells also revealed unexpected biology in the gut.
Results
Entero-hepato mTECs express gut and liver epithelial programs and are marked by Lypd8
To better understand the molecular regulation of mimetic cells, we performed detailed characterization of a mimetic cell type with transcriptomic features related to both gut enterocytes and liver hepatocytes, hereafter referred to as entero-hepato mTECs. Study of this subset was attractive for several reasons: (1) no prior work has examined the biology of entero-hepato mTECs in any detail; (2) we were able to develop protocols to isolate entero-hepato mTECs and characterize their molecular regulators using commercially available reagents; and (3) entero-hepato mTECs were putatively controlled by a family of lineage-defining TFs with only two members, simplifying generation of knockout mice in comparison with mimetic cell types putatively controlled by TF families with three or more members.
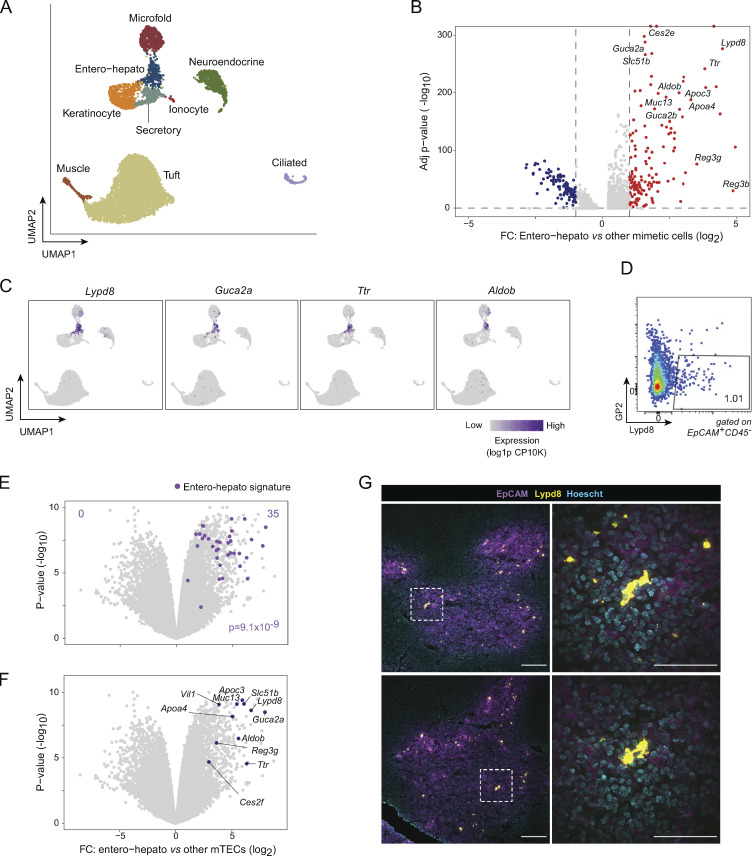
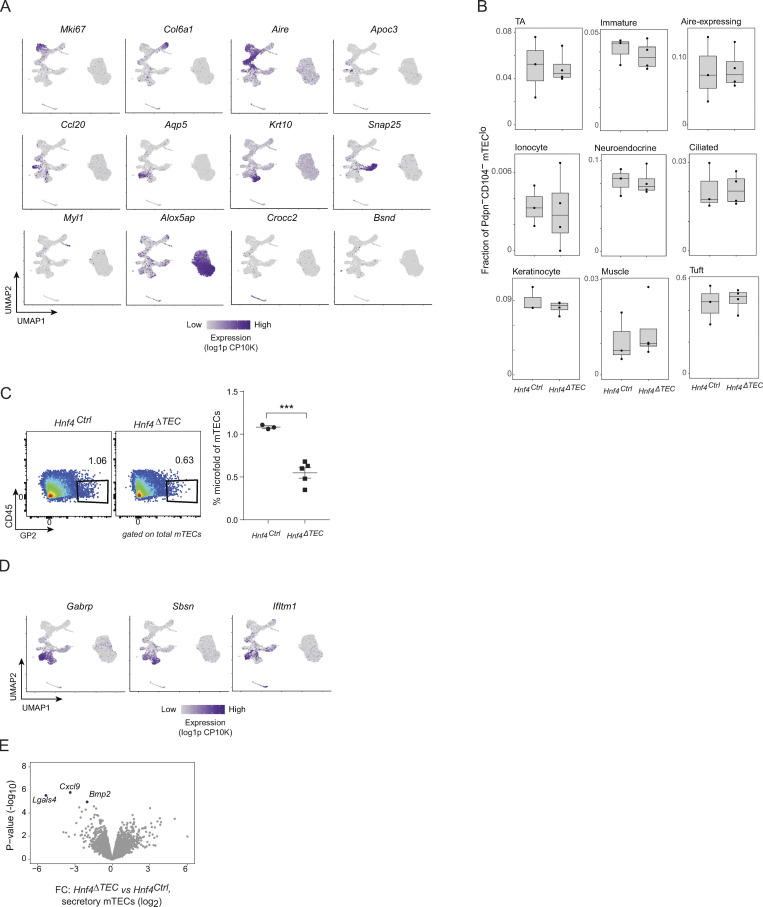
We began by closely examining the transcriptomic program of entero-hepato mTECs. Analyzing our previously published single-cell RNA sequencing (scRNA-seq) of mimetic cells (Michelson et al., 2022b), we observed a discrete cluster of entero-hepato mTECs, nearest in uniform manifold approximation and projection (UMAP) space to secretory (displaying mixed features of lung alveolar, club, and goblet cells), keratinocyte, and microfold mTECs (Fig. 1 A and Fig. S1 A). Analysis of differentially expressed transcripts by entero-hepato mTECs compared with the other mimetic cell types revealed that entero-hepato mTECs upregulated numerous transcripts encoding proteins associated with gut and liver epithelium, including apolipoproteins (Apoa4 and Apoc3), metabolic enzymes (Aldob and Ces2e), secreted peptide mediators (Guca2a and Guca2b), antimicrobial peptides (Reg3g, Reg3b, and Lypd8), bile acid transporters (Slc51b), serum transport proteins (Ttr), and mucins (Muc13; Fig. 1 B). Examination of the expression of a few of these transcripts in UMAP space confirmed their strong and specific expression in entero-hepato mTECs (Fig. 1 C). Thus, entero-hepato mTECs were a discrete subtype of mimetic cells that highly expressed gut- and liver-associated gene programs.
Figure 1.
Entero-hepato mTECs express gut and liver epithelial programs and are marked by Lypd8. (A) UMAP plot of scRNA-seq of mimetic cells, colored by mimetic cell cluster. Data were reanalyzed from Michelson et al. (2022b). For all UMAP plots, each dot represents a single cell. (B) Volcano plot of differentially expressed genes (highlighted red and blue, BH FDR <0.05) in entero-hepato mTECs vs. all other mimetic cells. For all volcano plots, each dot represents one gene. (C) UMAP plots as in A, colored by expression of the indicated transcripts. log1p CP10K, natural log1p of counts per 10,000 total counts. (D) Flow plot of entero-hepato mTECs (Lypd8+GP2−). (E) Volcano plot of bulk RNA-seq of entero-hepato mTECs (n = 3), gated as in D, versus other mTECs (Lypd8-GP2−; n = 3). The entero-hepato mTEC signature is overlaid in purple, and the P value was calculated by one-way Chi-squared test. (F) Data from E, with some entero-hepato marker genes labeled. (G) Immunofluorescence microscopy of thymic sections, stained for the indicated markers. Magnified views on the right are maximum intensity projections corresponding to boxed regions on the left. Scale bars, 100 μm (left) and 50 μm (right). For D and G, data are representative of at least two independent experiments.
Figure S1.
Additional characterization of entero-hepato mTECs. (A) UMAP plots of scRNA-seq of mimetic cells as in Fig. 1, colored by expression of indicated mimetic-cell genes. (B) Gating strategies for mTEC subsets. (C) Immunofluorescence microscopy of thymic section stained for Lypd8 and GP2. Scale bar, 100 μm. (D) UMAP plots of scATAC-seq of mimetic cells as in Fig. 2, colored by accessibility of the indicated mimetic-cell TF motifs. (E) Immunofluorescence microscopy of thymic section stained for Lypd8, Hnf4γ, and EpCAM. Scale bar on full image, 100 μm; scale bar on insets, 10 μm. Note that the full-size image corresponds to one of the sections shown in Fig. 1 G and Fig. 2 C. (F) Additional genome browser views of Hnf4α, Hnf4γ, and IgG CUT&Tag signal in mTECs at the indicated loci. Signal is in CPM. (G) Distribution of Hnf4γ and Aire peaks in mTECs at different genomic elements (top), and enrichment of each genomic element within said peaks (bottom). (H) Levels of Hnf4γ binding at entero-hepato OCRs, stratified by Aire co-binding. Signal is in CPM. For C and E, data are representative of at least two independent experiments. miRNA, microRNA; ncRNA, non-coding RNA; rRNA, ribosomal RNA; snoRNA, small nucleolar RNA; TTS, transcription termination site.
To permit rapid and routine study of entero-hepato mTECs, we devised a cytofluorimetric approach to isolate them. Lypd8, which encodes a glycosylphosphatidylinositol-linked antimicrobial peptide expressed on the surface of intestinal epithelial cells (Okumura et al., 2016), was among the most highly upregulated transcripts in entero-hepato mTECs (Fig. 1 B). By flow cytometry, we observed a clear Lypd8+ subset of mTECs, which comprised about 1% of the total mTEC pool after gating on GP2− cells to exclude microfold mTECs (Fig. 1 D and Fig. S1 B). To confirm that Lypd8+GP2− mTECs were entero-hepato mTECs, we performed population-level RNA-seq on purified Lypd8+GP2− mTECs versus Lypd8− mTECs. mTECs thus purified upregulated the entire entero-hepato mTEC signature, a set of 35 transcripts specifically associated with entero-hepato mTECs in our scRNA-seq data (Fig. 1 E). Focused examination of transcripts previously observed to be upregulated in entero-hepato mTECs by scRNA-seq, such as Aldob, Guca2a, and Slc51b, similarly found these transcripts to be upregulated in Lypd8+GP2− mTECs (Fig. 1 F). We concluded that entero-hepato mTECs could be purified cytofluorimetrically as Lypd8+GP2− mTECs.
Many mimetic cell types have spatially restricted patterns within the thymus, such as keratinocyte mTECs within Hassall’s corpuscles and ciliated mTECs within respiratory cysts (Hassall, 1846; Remak, 1855; Farr et al., 2002; Michelson et al., 2022b). We therefore sought to localize entero-hepato mTECs within the thymic epithelium. By immunofluorescence microscopy, we routinely observed bright and discrete Lypd8+ cells restricted to the EpCAM+ thymic medulla (Fig. 1 G). Individual Lypd8+ cells did not markedly cluster with one another within medullary islets. Lypd8 staining for entero-hepato mTECs was largely independent of GP2 staining for microfold mTECs (Fig. S1 C), though a few double-positive cells were observed, consistent with our cytofluorimetric results. Thus, entero-hepato mTECs populated the thymic medulla.
Hnf4 defines the chromatin program of entero-hepato mTECs
Given the general principle that lineage-defining TFs govern mimetic cell differentiation, we next sought to define the lineage-defining TF(s) controlling entero-hepato mTECs. In the periphery, enterocytes and hepatocytes both depend on the Hnf4 TF family, consisting of two homologs, Hnf4α and Hnf4γ, for their differentiation and function. In mice, enterocytes redundantly rely on Hnf4α and Hnf4γ, whereas hepatocytes strictly require Hnf4α but not Hnf4γ: mice lacking Hnf4a in the germline or specifically in liver epithelium die during development (Parviz et al., 2002; Parviz et al., 2003); mice lacking Hnf4a specifically in the intestine or Hnf4g in the germline are viable and reportedly healthy (Gerdin et al., 2006; Chen et al., 2019); and mice lacking Hnf4g in the germline that are then acutely deleted for Hnf4a specifically in the intestine die within short order (Chen et al., 2019).
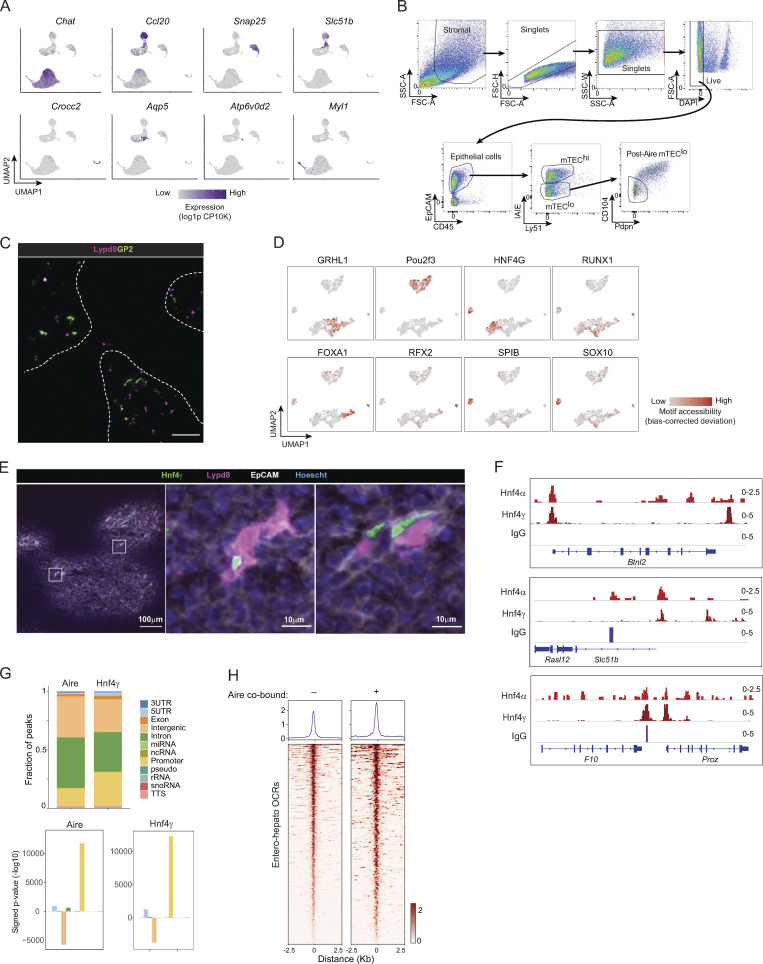
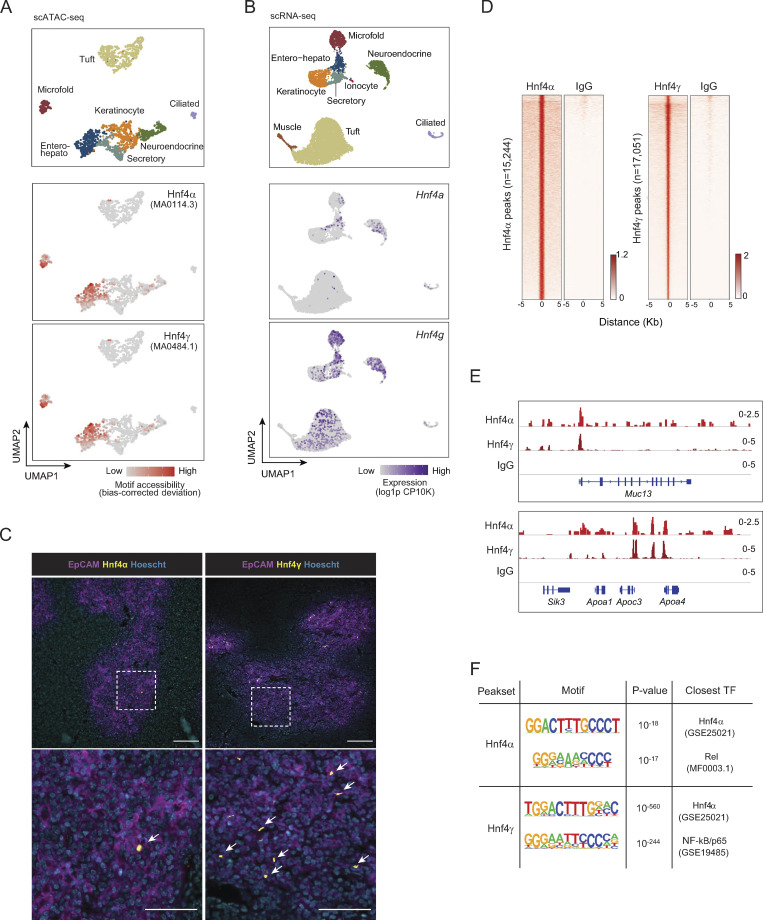
Motif analysis of our previously published single-cell assay for transposase-accessible chromatin with sequencing (scATAC-seq) data from mTECs revealed that Hnf4α and Hnf4γ had enriched motif accessibility within the accessible chromatin of both entero-hepato mTECs and microfold mTECs (Fig. 2 A and Fig. S1 D; Michelson et al., 2022b). The accessibility of Hnf4 motifs in microfold mTECs was biologically plausible, as microfold cells, which are specialized epithelial cells that flux luminal antigen into mucosa-associated lymphoid tissue, are derived from intestinal epithelial precursors in the gut (Gebert et al., 1999). Entero-hepato mTECs, however, were solely defined by Hnf4 motif accessibility, unlike microfold mTECs, which also harbored enhanced motif accessibility of the key microfold lineage-defining TFs, SpiB and Sox8 (Fig. S1 D). We therefore focused on Hnf4 family members as key regulators of entero-hepato mTECs.
Figure 2.
Hnf4 defines the chromatin program of entero-hepato mTECs. (A) UMAP plots of scATAC-seq of mimetic cells, reanalyzed from Michelson et al. (2022b). Cells are colored by mimetic cell type (top plot) or indicated motif accessibility (bottom two plots). (B) UMAP plots of scRNA-seq of mimetic cells, as in Fig. 1. Cells are colored by mimetic cell type (top plot) or indicated transcript expression (bottom two plots). (C) Immunofluorescence microscopy of thymic sections, stained for the indicated markers. Magnified views on the bottom are maximum intensity projections corresponding to boxed regions on the top. Scale bars, 100 μm (top) and 50 μm (bottom). Data are representative of at least two independent experiments. (D) Hnf4α and IgG CUT&Tag signal at Hnf4α peaks (left) and Hnf4γ and IgG CUT&Tag signal at Hnf4γ peaks (right) in purified mTECs. Signal is normalized as CPM. (E) Genome browser tracks of CUT&Tag of the indicated factors in mTECs at the indicated loci. Signal is in CPM. (F) De novo motifs (left) detected by HOMER within Hnf4α peaks (top) and Hnf4γ peaks (bottom), with corresponding P values for motif enrichment (center) and the name of the closest matched motif (right).
Returning to the scRNA-seq data, we noted that both Hnf4a and Hnf4g were expressed within entero-hepato mTECs, with Hnf4a more specifically but more weakly expressed within entero-hepato mTECs compared with Hnf4g, which was more strongly and more diffusely expressed (Fig. 2 B). Immunofluorescence microscopy of thymus sections confirmed Hnf4α and Hnf4γ expression, here localized to the nuclei of EpCAM+ epithelial cells within the thymic medulla (Fig. 2 C). Consistent with the scRNA-seq data, Hnf4α expression was relatively sparse, marking only a few cells per medullary islet, whereas Hnf4γ expression was more abundant. Many Hnf4γ+ cells were also Lypd8+ (Fig. S1 E).
To explore the integration of Hnf4α and Hnf4γ into mTEC chromatin landscapes, we mapped Hnf4γ binding to mTEC chromatin using cleavage under targets and tagmentation (CUT&Tag), a low-input technique for mapping chromatin-associated factors (Kaya-Okur et al., 2019), and jointly analyzed its binding with our previously generated CUT&Tag analysis of Hnf4α binding in mTECs (Michelson et al., 2022b). Our CUT&Tag data were of high quality, with good alignment to the Mus musculus genome over the spike-in Escherichia coli genome, adequate sequencing depth, and enrichment of signal in peaks (Table S1; Fig. 2, D and E; and Fig. S1 F). We detected similar numbers of binding peaks for the two factors on mTEC chromatin: 15,244 for Hnf4α and 17,051 for Hnf4γ (Fig. 2 D). Hnf4α and Hnf4γ both bound strongly to entero-hepato–associated loci, such as Muc13 and Apoa4/Apoc3/Apoa1 (Fig. 2 E and Fig. S1 F). The binding strength of Hnf4γ, as assessed by the CUT&Tag signal-to-noise ratio, appeared to be substantially stronger than that of Hnf4α, consistent with the more abundant expression of Hnf4γ versus Hnf4α in mTECs (Fig. 2, B and C). Motif analysis confirmed that the binding peaks of Hnf4α and Hnf4γ were both enriched for Hnf4 family motifs, again with more significant enrichment in Hnf4γ than Hnf4α peaks, consistent with specific activity of Hnf4α and Hnf4γ on mTEC chromatin at Hnf4 motifs (Fig. 2 F). Interestingly, there was also a substantial co-enrichment within Hnf4α and Hnf4γ binding peaks of NF-κB motifs (Rel and NF-κB/p65; Fig. 2 F), perhaps reflecting the prominent influence of receptor activator of NF-κB (RANK) signaling on mTEC biology (Rossi et al., 2007).
Finally, we examined the extent to which Hnf4 colocalized with Aire in mTECs using previously published Aire binding data (Bansal et al., 2017). Aire was co-bound at a large majority of Hnf4γ peaks (13,581 of 17,051 peaks; 79.6%). The types of genomic elements bound by the two factors were similar, with strong enrichment near genes and depletion from intergenic regions (Fig. S1 G). Hnf4γ peaks where Aire cobound had modestly higher levels of Hnf4γ than peaks lacking Aire, indicating that Aire was not required for basal Hnf4γ binding but that it could locally augment Hnf4γ levels (Fig. S1 H). These results were in line with previously described interactions between Aire and the lineage-defining TFs Grhl1 and Pou2f3 (Michelson et al., 2022b).
Comparative analysis of entero-hepato mTECs and bona fide enterocytes
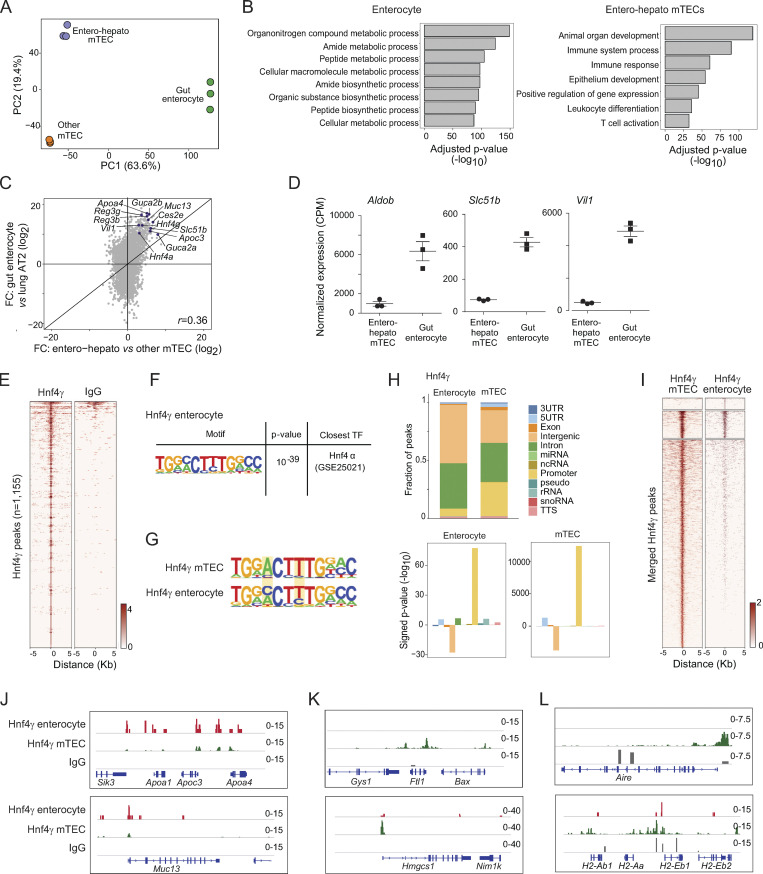
To evaluate the degree of molecular similarity between mimetic cells and their peripheral counterparts, we performed an in-depth comparison of entero-hepato mTECs and bona fide enterocytes. We began by comparing our RNA-seq of entero-hepato mTECs to RNA-seq of enterocytes previously prepared under identical library generation and sequencing conditions (Michelson et al., 2022b). Principal component analysis (PCA) of entero-hepato mTECs, all other mTECs (excluding entero-hepato mTECs), and bona fide enterocytes revealed that the major axis of transcriptional variation among these subsets (63.6% of the variance) separated thymic epithelium from peripheral epithelium, indicating that entero-hepato mTECs retained a core thymic identity (Fig. 3 A). Nonetheless, the second principal component, comprising a minor but substantial 19.4% of the variance, separated entero-hepato mTECs from other mTECs, indicating that entero-hepato mTECs layered on a substantial mimetic-cell-specific transcriptomic program (Fig. 3 A). Differential expression of entero-hepato mTECs and enterocytes found 3,513 transcripts upregulated in enterocytes and 6,782 transcripts in entero-hepato mTECs (fold-change [FC] >2, false discovery rate [FDR] <0.05), enriched for nutrient metabolic and biosynthetic pathways in enterocytes and immune system and epithelial development pathways in entero-hepato mTECs (Fig. 3 B).
Figure 3.
Comparative genomic analysis of entero-hepato mTECs and bona fide enterocytes. (A) PCA plot of bulk RNA-seq of entero-hepato mTECs, other mTECs, and bona fide enterocytes. Each dot represents one biological replicate. (B) Gene ontology plots of differentially upregulated genes in entero-hepato mTECs and enterocytes. (C) FC/FC plot of bulk RNA-seq of entero-hepato mTECs versus other mTECs on the x-axis and bona fide enterocytes versus lung alveolar epithelial cells (AT2 cells) on the y-axis, with some enterocyte marker genes labeled. Each dot represents one gene. (D) Normalized expression of the indicated enterocyte marker genes in entero-hepato mTECs and enterocytes. Each dot represents one biological replicate and bars represent mean ± SEM. (E) Hnf4γ CUT&Tag signal at Hnf4γ peaks in enterocytes, compared with IgG. (F) De novo motif (left) detected by HOMER within Hnf4γ peaks, with corresponding P value for motif enrichment (center) and the name of the closest matched motif (right). (G) Comparison of motif sequence logos for Hnf4γ peaks in mTECs (top) and enterocytes (bottom). Key areas of difference are highlighted. (H) Distribution of Hnf4γ peaks in mTECs and enterocytes at different genomic elements (top), and enrichment of each genomic element within said peaks (bottom). (I) Comparison of Hnf4γ binding in enterocytes and mTECs at merged enterocyte and mTEC Hnf4γ peaks. Peaks are stratified by k-means clustering (k = 3). (J–L) Genome browser tracks for the indicated factors at loci containing the indicated enterocyte and mTEC marker genes. For E and I–L, signal is in CPM. miRNA, microRNA; ncRNA, non-coding RNA; rRNA, ribosomal RNA; snoRNA, small nucleolar RNA; TTS, transcription termination site.
FC/FC analysis permitted direct comparison of transcripts upregulated in entero-hepato mTECs vis-à-vis the other mTECs and in bona fide enterocytes vis-à-vis lung alveolar epithelial cells (employed here as an epithelial outgroup; Fig. 3 C). Overall, the FCs were moderately well correlated (r = 0.36, P < 0.0001). Examination of the FC/FC plot yielded three major conclusions: first, substantial numbers of transcripts were upregulated in entero-hepato mTECs and bona fide enterocytes; second, the transcripts most highly upregulated in entero-hepato mTECs were in general also the transcripts most highly upregulated in enterocytes; and third, the strength of upregulation of these shared transcripts was several orders of magnitude higher in enterocytes than in entero-hepato mTECs (Fig. 3 C). Many of these shared upregulated transcripts were the usual suspects from our close analysis of entero-hepato mTECs (i.e., Fig. 1, B and F), such as Apoa4, Reg3g, and Muc13. Direct analysis of jointly normalized expression values of several enterocyte-specific transcripts confirmed their transcription in entero-hepato mTECs but with an order-of-magnitude higher level of expression in bona fide enterocytes (Fig. 3 D). Altogether, we concluded that while entero-hepato mTECs retained a core thymic gene program, they simultaneously upregulated a wide swath of the bona fide enterocyte gene program. The magnitude of this upregulation was smaller than that of bona fide enterocytes but the breadth of transcripts induced was nonetheless surprisingly comprehensive.
To understand how the activity of Hnf4 in mTECs compared with its activity in enterocytes, we generated CUT&Tag of Hnf4γ in enterocytes using identical library preparation techniques as for mTECs. We focused on Hnf4γ given its strong binding enrichment in the thymus relative to Hnf4α and because Hnf4α and Hnf4γ bind nearly identically in the gut (Chen et al., 2019). Our CUT&Tag data were again of good quality (Table S1). We could detect Hnf4γ binding over background at 1,155 peaks in enterocytes (Fig. 3 E). Motif analysis confirmed that these peaks were enriched for Hnf4 family motifs (Fig. 3 F). Comparison of the Hnf4γ de novo binding motif in thymus versus intestine revealed a broadly similar motif selectivity, but some variations were apparent, including a preference for adenosine at position 4 and increased stringency for thymidine at position 7 (Fig. 3 G). Unlike in the thymus, NF-κB family motifs were not appreciably enriched within intestinal Hnf4γ peaks, which may be reflected in the variation in organ-specific Hnf4γ motif preference. No other notable TF motif differences in thymus and gut Hnf4γ binding sites were observed. In both the thymus and gut, Hnf4γ binding was enriched near genes (i.e., promoters and enhancers) and depleted in intergenic regions, with a somewhat higher fraction of promoters bound in the thymus relative to the gut (Fig. 3 H).
Joint analysis of thymic and intestinal Hnf4γ peaks revealed three classes of Hnf4γ binding, corresponding to enterocyte-specific, shared, and mTEC-specific sites (Fig. 3 I). At loci encoding enterocyte-expressed transcripts like Apoa4 and Muc13, the relative strength of Hnf4γ binding was stronger in enterocytes than in mTECs, perhaps reflecting a greater amount of TF required to maximize transcriptional output of these key functional molecules in enterocytes (Fig. 3 J). At another set of loci, encoding transcripts specifying metabolic and serum proteins more typically associated with hepatocytes, such as Ftl1, encoding ferritin light chain, and Hmgcs1, encoding the cholesterol-synthetic enzyme HMG-CoA synthase, the inverse was true, with stronger Hnf4γ binding in mTECs and weaker binding in enterocytes (Fig. 3 K). Finally, at a set of loci with gene products associated with mTECs, such as Aire and the major histocompatibility complex class II (MHCII), we observed strong Hnf4γ binding in mTECs but generally less binding in enterocytes, though at some sites, such as within the MHCII locus, Hnf4γ also was bound in enterocytes, consistent with their known ability to upregulate MHCII molecules (Fig. 3 L; Bland and Warren, 1986). Thus, Hnf4γ bound loci encoding enterocyte-specific gene products more strongly in enterocytes than mTECs but bound more diverse loci in mTECs than enterocytes, including loci encoding hepatocyte- and mTEC-specific gene products.
Hnf4 is required for entero-hepato mTEC accumulation and PTA expression
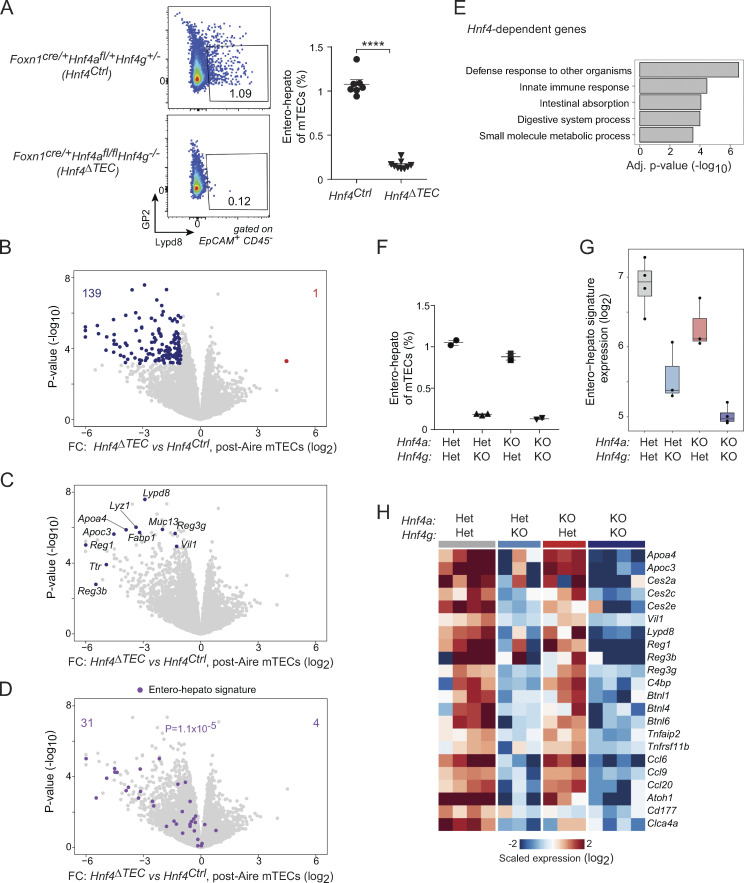
We next asked whether Hnf4, given its central role in the chromatin program of entero-hepato mTECs, was required for entero-hepato mTEC accumulation and/or PTA expression. To this end, we generated triple-transgenic mice lacking both Hnf4α and Hnf4γ in TECs by crossing together Foxn1cre, Hnf4aflox, and Hnf4g-null genetic elements on the C57BL/6J (B6) genetic background. Control animals were of the genotype Foxn1cre/+Hnf4afl/+Hnf4g+/− (hereafter Hnf4Ctrl), while experimental animals were of the genotype Foxn1cre/+Hnf4afl/flHnf4g−/− (hereafter Hnf4ΔTEC), yielding mice that were conditionally deleted for Hnf4α in Foxn1-expressing TECs and germline-deleted for Hnf4γ. Offspring from these crosses were viable and born at normal Mendelian ratios. Hnf4ΔTEC mice were moderately smaller than their littermate controls, but the relative distributions of major TEC and thymocyte subtypes were not altered (Fig. S2, A–F).
Figure S2.
Thymic compartments in Hnf4ΔTEC mice. (A) Body weights of Hnf4Ctrl (n = 8) versus Hnf4ΔTEC (n = 4) mice. (B) Thymus weights of Hnf4Ctrl (n = 8) versus Hnf4ΔTEC (n = 4) mice. (C) Thymic epithelial cell (EpCAM+ CD45−) numbers in Hnf4Ctrl (n = 3) versus Hnf4ΔTEC (n = 6) mice. (D) Fractional abundance of the indicated TEC compartments in Hnf4Ctrl (n = 6) versus Hnf4ΔTEC (n = 3) mice. (E) Hematopoietic cell (CD45+) numbers in Hnf4Ctrl (n = 8) versus Hnf4ΔTEC (n = 5) mice. (F) Fractional abundance of the indicated thymocyte compartments in Hnf4Ctrl (n = 8) versus Hnf4ΔTEC (n = 4) mice. For A, B, and D–F, data were pooled from two independent experiments. For C, data are representative of at least two independent experiments. (G) Expression of Aire in MHCII-high mTECs from Hnf4Ctrl and Hnf4ΔTEC mice, assayed by bulk RNA-seq. (H) Expression of Hnf4a and Hnf4g in MHCII-high mTECs from Aire+/+ and Aire−/−, assayed by bulk RNA-seq reanalyzed from Bansal et al. (2021). For all plots, each dot represents one mouse and bars represent mean ± SEM.
We first measured the abundance of entero-hepato mTECs by flow cytometry. Whereas Lypd8+GP2− entero-hepato mTECs comprised about 1% of the total mTEC pool in Hnf4Ctrl mice, similar to wildtype mice, entero-hepato mTECs were nearly totally ablated in Hnf4ΔTEC mice (Fig. 4 A). Thus, Hnf4 was required for entero-hepato mTEC accumulation. Population-level RNA-seq of post-Aire mTECs from Hnf4Ctrl and Hnf4ΔTEC mice revealed that, in the absence of Hnf4, 139 transcripts were significantly downregulated (FDR < 0.05) and only one transcript significantly upregulated (Fig. 4 B). These transcripts included many of the previously noted entero-hepato mTEC marker transcripts, including Apoa4, Muc13, Ttr, and Reg3g (Fig. 4 C). Overlaying the entero-hepato mTEC signature derived previously confirmed that nearly all of the signature was downregulated in the absence of Hnf4 (Fig. 4 D). Pathway analysis showed that the downregulated transcripts were enriched for enterocyte/hepatocyte-specific biological pathways, including antimicrobial, digestive, and metabolic processes (Fig. 4 E). In sum, mTECs specifically downregulated PTA expression associated with entero-hepato mTECs in the absence of Hnf4. Notably, Aire expression was unchanged in MHCII-high mTECs (mTEChi) from Hnf4ΔTEC mice (Fig. S2 G).
Figure 4.
Hnf4 is required for entero-hepato mTEC accumulation and PTA expression. (A) Representative flow plots (left) and summarized data (right) of the fraction of entero-hepato mTECs in thymi from Hnf4Ctrl (n = 7) versus Hnf4ΔTEC (n = 9) mice. For summarized data, each dot represents one mouse, bars represent mean ± SEM, data were pooled from three independent experiments, and P value was calculated by two-sided, unpaired Student’s t test. ****, P < 0.0001. (B–D) Volcano plots of bulk RNA-seq of post-Aire (Pdpn−CD104−) mTECs from Hnf4Ctrl (n = 4) versus Hnf4ΔTEC (n = 4) mice, colored by differentially expressed genes (top; BH FDR < 0.05), selected entero-hepato markers (middle), or the entero-hepato mTEC signature (bottom; P value calculated by one-way Chi-squared test). (E) Several top gene ontology terms among the transcripts significantly downregulated in post-Aire mTECs from Hnf4ΔTEC mice, as indicated in B. Enrichment was calculated using gProfiler. (F) Fractional abundance of entero-hepato mTECs in mice with the indicated genotypes. Each dot represents one mouse, bars represent mean ± SEM, and data for 4aHet4gHet and 4aKO4gKO mice were replotted from A. Data are representative of at least two independent experiments. (G) Mean expression of entero-hepato mTEC signature genes in bulk RNA-seq of post-Aire mTECs from 4aHet4gHet (n = 4), 4aHet4gKO (n = 3), 4aKO4gHet (n = 3), and 4aKO4gKO (n = 4) mice. For all boxplots, median and interquartile range (IQR) are shown as boxes and minimum and maximum values (up to ±1.5*IQR) as whiskers. (H) Heatmap of scaled expression of select entero-hepato mTEC marker genes in post-Aire mTECs from mice of the indicated genotypes. Each row is one gene, and each column is one biological replicate.
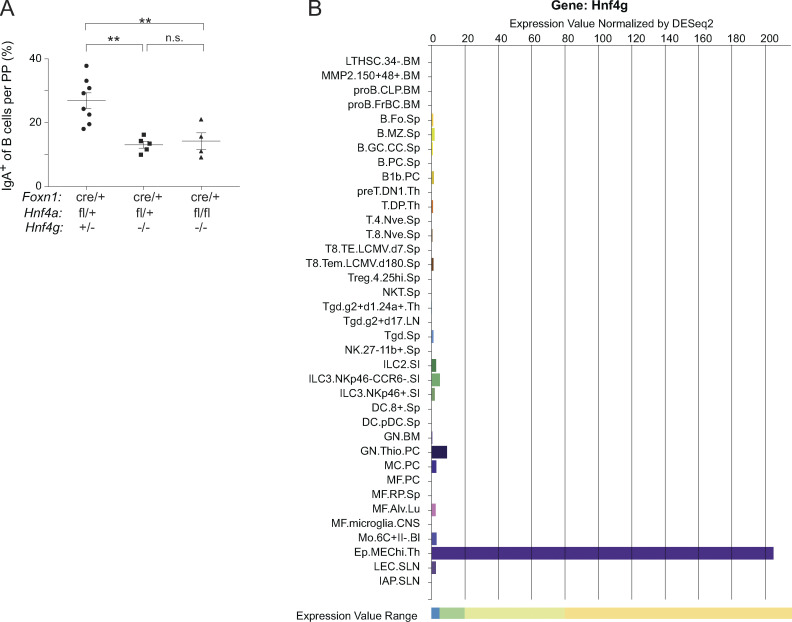
As we deleted both Hnf4α and Hnf4γ in mTECs, these effects might have reflected the actions of solely one or the other, or some combination of the two. We therefore analyzed entero-hepato mTECs in Foxn1cre/+Hnf4afl/+Hnf4g+/− (4aHet4gHet), Foxn1cre/+Hnf4afl/+Hnf4g−/− (4aHet4gKO), Foxn1cre/+Hnf4afl/flHnf4g+/− (4aKO4gHet), and Foxn1cre/+Hnf4afl/flHnf4g−/− (4aKO4gKO) mice to understand the relative contribution of each factor to entero-hepato mTEC accumulation and PTA expression. Cytofluorimetric analysis of entero-hepato mTECs showed that they were again ablated in 4aHet4gKO mice but were largely preserved in 4aKO4gHet mice, indicating that, unlike in most other organs (Parviz et al., 2003; Chen et al., 2019), Hnf4γ was the major Hnf4 family TF required for entero-hepato mTEC accumulation (Fig. 4 F). Similarly, population-level RNA-seq of post-Aire mTECs from the four genotypes showed a substantial downregulation of the entero-hepato mTEC signature in 4aHet4gKO mice relative to 4aHet4gHet controls, but relatively little downregulation in 4aKO4gHet mice (Fig. 4 G). Notably, however, the entero-hepato mTEC signature was most substantially downregulated in post-Aire mTECs from 4aKO4gKO mice relative to either single deletion, and even in 4aKO4gHet mice, there was a trend toward diminished expression of the entero-hepato mTEC signature, suggesting that, while Hnf4γ was the major factor required, Hnf4α played a minor but not fully redundant role in promoting entero-hepato mTEC accumulation and PTA expression. Direct analysis of the expression of transcripts associated with entero-hepato mTECs confirmed that the major trend for Hnf4 deletion followed the Hnf4g genotype, but that a few genes, especially enteroendocrine-associated genes like Atoh1 and Cd177, required Hnf4α for their full expression (Fig. 4 H). Interestingly, analysis of previously published RNA-seq data of mTECs from Aire−/− mice showed that Hnf4g expression was essentially unchanged in the absence of Aire whereas Hnf4a expression was nearly totally ablated (Fig. S2 H; Bansal et al., 2021). This result was consistent with our previous finding that entero-hepato mTEC abundance and cell state are largely independent of Aire (Michelson et al., 2022b), but also suggested that Aire may influence the Hnf4α-driven component of PTA expression.
Overall, we concluded that Hnf4-family TFs were required for the accumulation of entero-hepato mTECs and expression of their associated PTAs. Of the two Hnf4 family members, Hnf4γ played the major role, but Hnf4α also appeared to have a minor, non-redundant contribution.
Hnf4 activates enhancers and redistributes CTCF but does not impact promoter state or Polycomb-mediated repression
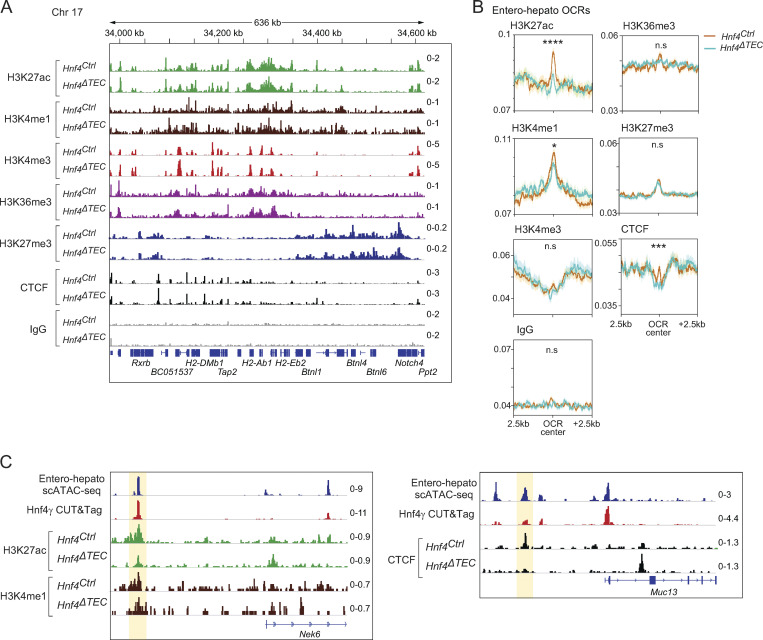
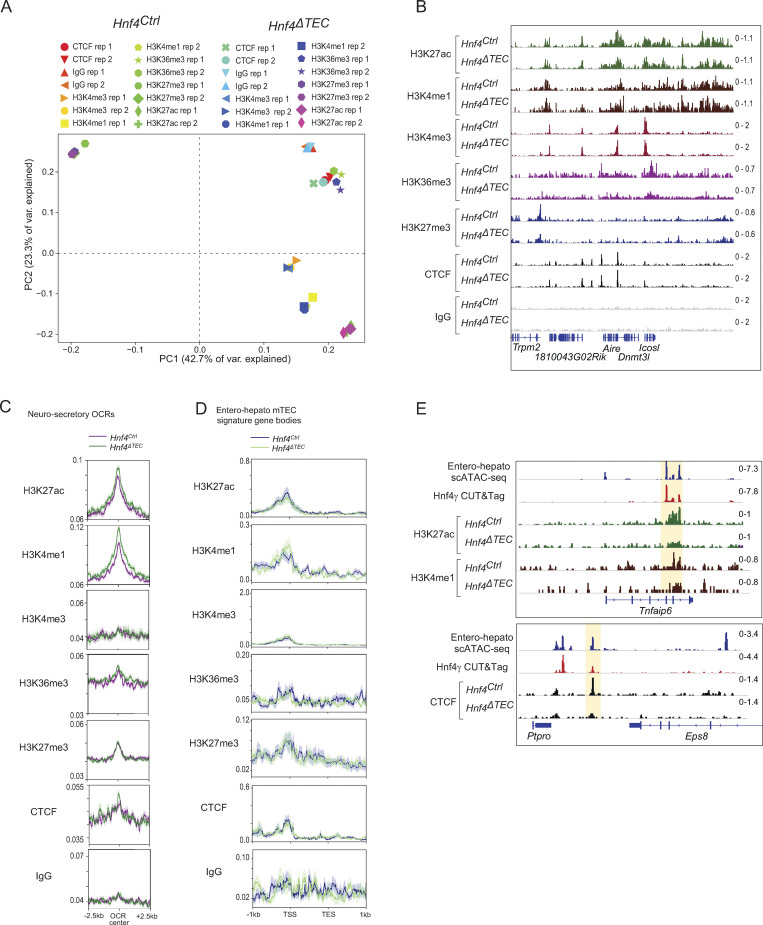
To gain a molecular understanding of how Hnf4-family TFs promoted entero-hepato mTEC differentiation and function, we performed extensive chromatin profiling of mTECs from Hnf4Ctrl and Hnf4ΔTEC mice, generating cleavage under targets and release under nuclease (CUT&RUN) profiles for seven different histone marks or chromatin-binding proteins (H3K27ac, H3K4me1, H3K4me3, H3K36me3, H3K27me3, and CTCF, plus IgG as a control) in duplicate for both genotypes. The CUT&RUN data were of good quality, with good enrichment of the M. musculus genome over the spike-in E. coli genome, adequate sequencing depth, enriched signal within peaks, and as-expected clustering by chromatin factor and biological replicate upon PCA of binned genomic signal (Table S1; Fig. 5 A; and Fig. S3 A).
Figure 5.
Hnf4 acts on enhancers and CTCF, but not promoters or heterochromatin. (A) Genome browser tracks for merged CUT&RUN signal (n = 2 replicates for each) for the indicated factors in mTECs derived from mice of the indicated genotypes at the H2-A/E loci. Signal is in CPM. (B) Profile plots of aggregated signal for the indicated factors centered on entero-hepato–specific OCRs plus 2.5 kilobases (kb) upstream and downstream. Signal is in CPM, dark lines indicate mean signal, and shading around lines indicates SEM. P-values were calculated by two-sided, paired Student’s t test of CUT&RUN signal at the central 100 bp of entero-hepato OCRs in Hnf4Ctrl vs. Hnf4ΔTEC mice, with Bonferroni correction. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. (C) Genome browser tracks for the indicated factors in mTECs at the indicated loci. Signal is in CPM.
Figure S3.
Quality control of CUT&RUN in mTECs. (A) PCA plot of binned genomic signal in individual CUT&RUN replicates for all factors assayed in Hnf4Ctrl and Hnf4ΔTEC mice. (B) Genome browser tracks for the indicated factors at the Aire locus in mTECs from Hnf4Ctrl and Hnf4ΔTEC mice. (C) Profile plots of aggregated signal for the indicated factors centered on neurosecretory-specific OCRs plus 2.5 kb upstream and downstream. (D) Profile plots of aggregated signal for each genotype at the gene bodies of entero-hepato mTEC signature genes, aligned from transcription start site (TSS) to transcription end site (TES) plus 1 kb upstream and downstream. For C and D, dark lines represent the mean signal, shaded areas around dark lines represent SEM, and the signal is in CPM. (E) Genome browser tracks for the indicated factors in mTECs at the indicated loci. For B–E, the signal is in CPM.
Examination of the genome tracks for the different factors showed that each one had a distinct distribution of binding sites on mTEC chromatin, in patterns that agreed with our understanding of mTEC biology (Fig. 5 A). For instance, the H2-Aa/b and H2-Ea/b loci, encoding MHCII molecules, showed enrichment of H3K27ac, marking active enhancers and promoters, throughout, with H3K4me1 and H3K4me3 differentially marking subsets of active enhancers and active promoters, respectively. H3K36me3 was enriched in active gene bodies. In contrast, H3K27me3 was depleted from the active MHC-molecule-encoding loci but was enriched in the H3K27ac-poor neighboring Btnl locus encoding butyrophilin-like molecules associated with peripheral epithelia. Finally, CTCF had its own sharp, distinct binding peaks, consistent with its unique role in structuring the genome. There was little background detected in the IgG genome tracks. At a high level, the genome tracks for each factor were similar for mTECs derived from Hnf4Ctrl versus Hnf4ΔTEC mice, indicating that Hnf4 deletion did not induce a broad-scale rearrangement of the mTEC chromatin state. Nor did Hnf4 loss substantially impact chromatin state at the Aire locus (Fig. S3 B).
We therefore proceeded with focused analysis of Hnf4’s influence on chromatin factors at entero-hepato-specific open-chromatin regions (OCRs) defined by our scATAC-seq data (Fig. 5 B). Unlike the coarse-grained examination of genome tracks, this focused approach revealed a substantial induction of H3K27ac at entero-hepato OCRs in the presence of Hnf4, corresponding to an activation of enhancers and/or promoters. No such induction of H3K27ac—and in fact the reverse, a reduction in H3K27ac—was seen at OCRs associated with other mimetic cells (neurosecretory mTECs), indicating that this effect was specific to entero-hepato OCRs (Fig. S3 C). To tease apart whether this activation was an enhancer- or promoter-driven effect, we examined levels of H3K4me1 (marking active enhancers) and H3K4me3 (marking active promoters) at the same sites. H3K4me1 levels were enhanced at entero-hepato OCRs in the presence of Hnf4, albeit more modestly than was H3K27ac and, again, inversely reduced at neurosecretory OCRs (Fig. 5 B and Fig. S3 C). In contrast, H3K4me3 levels were essentially unchanged at both entero-hepato OCRs and neurosecretory OCRs in the presence or absence of Hnf4 (Fig. 5 B and Fig. S3 C). Thus, the enriched deposition of H3K27ac and H3K4me1, but not H3K4me3, at entero-hepato OCRs reflected increased activation of enhancers, but not promoters, by Hnf4. A similar mechanistic pattern has been described for Hnf4 in the gut (Chen et al., 2019).
H3K36me3 levels, marking active gene bodies, were not substantially changed at entero-hepato OCRs nor at the gene bodies of entero-hepato mTEC signature genes, consistent with the finding that Hnf4 activated enhancers but not promoters and suggesting that mimetic-cell PTA gene promoters were “poised” for expression regardless of the presence of their activating TFs (Fig. 5 B and Fig. S3 D). Along the same lines, examination of the distribution of all profiled chromatin factors at the promoters and gene bodies of entero-hepato mTEC signature genes revealed few Hnf4-dependent differences (Fig. S3 D). H3K27me3 levels were also very similar at entero-hepato OCRs regardless of the Hnf4 status, indicating that Hnf4 did not affect Polycomb-mediated repression at these sites (Fig. 5 B). Finally, CTCF levels were markedly enriched exactly at the center of entero-hepato OCRs, with an inverse effect observed at neurosecretory OCRs (Fig. 5 B and Fig. S3 D). Thus, Hnf4 promoted the deposition of CTCF at entero-hepato accessible regions, implying that Hnf4-driven architectural changes in mTEC genome structure were involved in the generation of entero-hepato mTECs. Again, little background was observed in the IgG traces (Fig. 5 B).
In sum, chromatin profiling of mTECs from Hnf4Ctrl and Hnf4ΔTEC mice showed that Hnf4 promoted activation of enhancers and CTCF deposition at entero-hepato OCRs. A few examples of these effects are shown in Fig. 5 C and Fig. S3 E. In contrast, few differences were observed in the activation of promoters or gene bodies or in Polycomb-mediated repression at the same sites or entero-hepato mTEC signature genes.
Hnf4 affects transcription in mTECs in three discernable manners
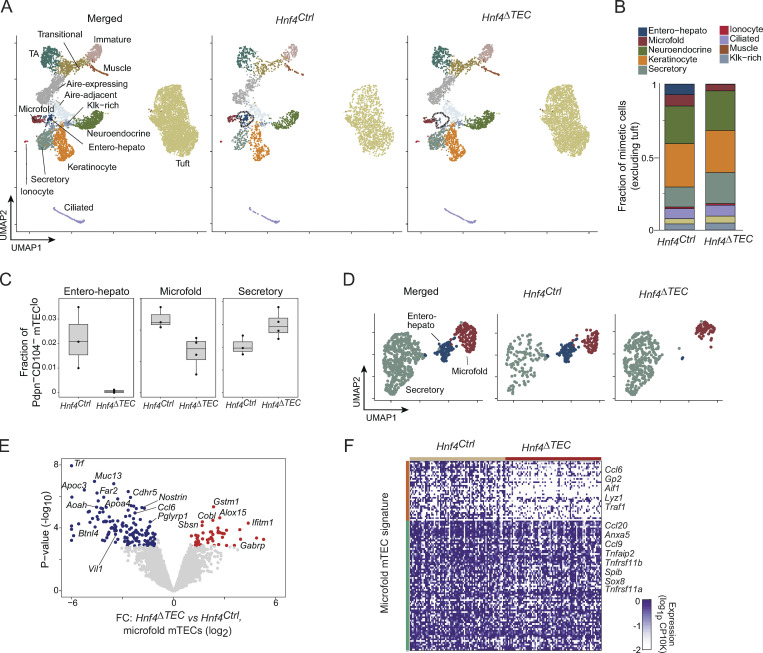
We next sought to connect the molecular changes in mTEC chromatin induced by Hnf4 to a high-resolution understanding of its transcriptional effects in the mTEC compartment. We performed scRNA-seq of mimetic cells from Hnf4Ctrl (n = 3) and Hnf4ΔTEC (n = 4) mice, hashtagging cells with barcoded antibodies to permit simultaneous sample preparation and eliminate batch effects (Stoeckius et al., 2018). After filtering, we retained 9,185 cells for visualization and downstream analysis, 3,295 from Hnf4Ctrl and 5,890 from Hnf4ΔTEC mice (Fig. 6 A). We could readily detect the various mimetic cell types (Fig. S4 A). Between the Hnf4Ctrl and Hnf4ΔTEC conditions, consistent with our cytofluorimetric analysis, we observed a near-total ablation of the entero-hepato mTEC cluster in the Hnf4ΔTEC condition (Fig. 6 A, dashed lines). The vast majority of other clusters were visually undisturbed, consistent with the idea that specific lineage-defining TFs are required only for their respective mimetic cell types.
Figure 6.
Three different effects of Hnf4 on mimetic cells. (A) UMAP plots of scRNA-seq of mimetic cells from Hnf4Ctrl (n = 3) and Hnf4ΔTEC (n = 4) mice, merged (left) or divided by genotype (center and right) and colored by cluster. (B) Stacked barplot indicating the mean fractional representation of each mimetic cell type in thymi from Hnf4Ctrl and Hnf4ΔTEC mice. Note that tuft mTECs, being disproportionately abundant, were excluded to allow for adequate visualization of other clusters. (C) Boxplots of fractional abundance of the indicated mimetic cell clusters among all post-Aire (Pdpn−CD104−) mTEC. Each dot represents one biological replicate. (D) Subclustered scRNA-seq UMAP plots of secretory, entero-hepato, and microfold mTECs from Hnf4Ctrl and Hnf4ΔTEC mice, merged (left) or divided by genotype (center and right) and colored by cluster. (E) Volcano plot of pseudobulked differential expression of microfold mTECs derived from Hnf4Ctrl and Hnf4ΔTEC mice. Differentially expressed genes (BH FDR < 0.05) are highlighted. (F) Heatmap of expression of microfold mTEC signature genes among microfold mTECs derived from Hnf4Ctrl and Hnf4ΔTEC mice. Each column is one cell, each row is one gene, cells are organized by genotype, and genes are k-means clustered (k = 2) with some representative genes for each cluster labeled.
Figure S4.
Additional analysis of scRNA-seq of mimetic cells from Hnf4Ctrl and Hnf4ΔTEC mice. (A) UMAP plots of scRNA-seq of mimetic cells from Hnf4Ctrl and Hnf4ΔTEC mice, colored by expression of the indicated mimetic-cell marker genes. (B) Boxplots of relative abundance of the indicated mimetic cell types among all Pdpn−CD104− mTEClo in individual scRNA-seq replicates, divided by genotype. Each dot represents one mouse. (C) Representative flow cytometry plots (left) and summarized data (right) of the relative abundance of microfold mTECs in thymi from Hnf4Ctrl (n = 3) and Hnf4ΔTEC (n = 5) mice. Each dot represents one mouse, bars show mean ± SEM, and data are representative of two independent experiments. ***, P < 0.001. (D) UMAP plots as in A, colored by expression of the indicated genes. (E) Volcano plot of pseudobulked differential expression of secretory mTECs derived from Hnf4Ctrl and Hnf4ΔTEC mice. Differentially expressed genes (BH FDR < 0.05) are highlighted.
Quantitation of relative cluster abundance among mimetic cells (excluding tuft mTECs due to that cluster’s large size) confirmed that entero-hepato mTECs were ablated but most other clusters were not affected by Hnf4 deletion in TECs (Fig. 6 B). However, two mimetic cell types—microfold mTECs and secretory mTECs—did show asymmetric changes in their relative abundances among mimetic cells, with microfold mTECs reduced by roughly half and secretory mTECs increased by about 50%. These effects were verified by direct quantification of the abundance of these clusters in individual scRNA-seq replicates (Fig. 6 C and Fig. S4 B) and by flow cytometric verification of the reduction in microfold mTECs (Fig. S4 C). We did not confirm the change in secretory mTEC abundance by flow cytometry because we currently lack a suitable staining panel to detect these cells.
To gain insight into the nature of these changes, we subclustered and revisualized entero-hepato, microfold, and secretory mTECs separately from the larger dataset (Fig. 6 D). Multiple Hnf4-dependent effects were apparent. First, entero-hepato mTECs simply disappeared in the absence of Hnf4. Second, secretory mTECs increased in fractional abundance but displayed a similar transcriptional state in the absence of Hnf4, as judged by their distribution in UMAP space. And third, microfold mTECs were present in Hnf4ΔTEC mice, albeit fractionally reduced in abundance, but shifted their transcriptional program such that their distribution in UMAP space was almost entirely non-overlapping with microfold mTECs from Hnf4Ctrl mice.
We directly compared the transcriptomes of microfold mTECs from Hnf4ΔTEC and Hnf4Ctrl mice by performing differential expression analysis of pseudobulk-aggregated microfold mTECs from each biological replicate, taking this approach to avoid the inflated false discoveries that can result from cell-level differential expression (Squair et al., 2021). We observed that, consistent with our visual observations in UMAP space, many transcripts were downregulated and upregulated in Hnf4-deficient microfold mTECs (Fig. 6 E). The downregulated transcripts, more numerous, tended to encode gut-associated molecules also expressed in entero-hepato mTECs, such as Apoc3, Muc13, and Vil1. Several transcripts encoding functional molecules specific to microfold cells, like Ccl6 and Pglyrp1, were downregulated as well. The upregulated transcripts, fewer in number, were more of a mixed group, including several transcripts encoding molecules also expressed by secretory mTECs, such as Sbsn, Gabrp, and Ifitm1 (Fig. S4 D). The same differential expression procedure applied to secretory mTECs showed that, by contrast, only three genes were differentially expressed in secretory mTECs from Hnf4ΔTEC versus Hnf4Ctrl mice, indicating largely similar (though not perfectly identical) transcriptional profiles (Fig. S4 E). Focused analysis of expression of the microfold mTEC gene signature in microfold mTECs from Hnf4ΔTEC versus Hnf4Ctrl mice showed that roughly one-third of the microfold signature was downregulated in the absence of Hnf4 (Fig. 6 F), including some transcripts encoding key microfold molecules, like Gp2 and Ccl6, but sparing others, like Spib, Sox8, Ccl9, and Ccl20.
Overall, then, we observed three distinct genres of transcriptional effects driven by Hnf4. For entero-hepato mTECs, loss of Hnf4 precluded generation of the mimetic cell type altogether. For secretory mTECs, loss of Hnf4 actually increased their relative abundance but did not substantially alter their transcriptional state, perhaps pointing toward a bifurcating fate choice between entero-hepato and secretory mTEC differentiation. Notably, this bias toward secretory lineages in the absence of Hnf4 was remarkably similar to the shift away from the enterocyte lineage and toward the intestinal secretory lineages seen when Hnf4 is acutely lost in intestinal epithelial progenitors (Chen et al., 2019). Finally, for microfold mTECs, loss of Hnf4 transformed their transcriptomes without ablating the subtype altogether, indicating that Hnf4 was required for a discrete Hnf4-dependent gene program in microfold mTECs but was nonessential for some basal accumulation of the subtype.
A requirement for Hnf4γ in gut microfold cells
Next, we wondered if we could learn new peripheral biology from mimetic cells. Many mimetic cell types quite faithfully mirror the known biology of their peripheral counterparts (Michelson et al., 2022b); we reasoned that certain as-yet unappreciated aspects of peripheral cell biology might also be mirrored in mimetic cell biology.
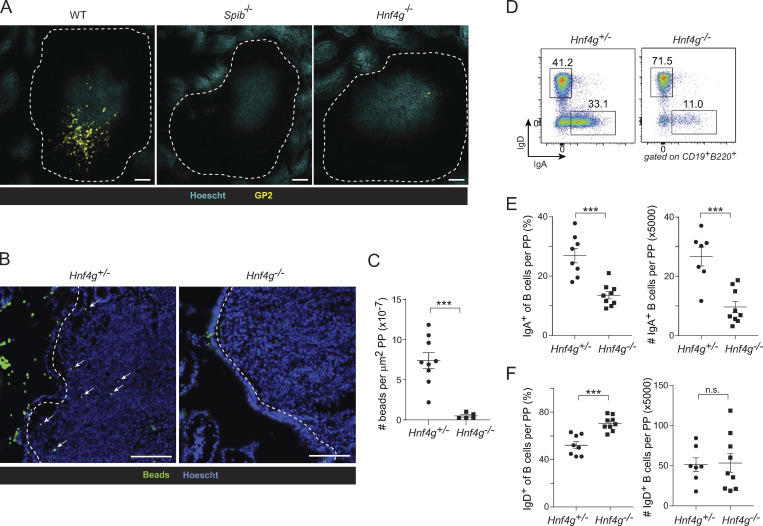
As a proof of principle, we focused on the microfold mTEC, whose peripheral counterpart, the gut microfold cell, plays important roles in the sampling of gut antigens and generation of gut immune responses (reviewed in Mabbott et al., 2013). Our data showed that Hnf4 controlled part of the gene program of microfold mTECs, but to our knowledge, there has been no corresponding implication of Hnf4 in the biology of gut microfold cells. We therefore examined our Hnf4ΔTEC mice, which were germline-deficient in Hnf4g, for defects in gut microfold cell accumulation and/or function. Remarkably, in the germline absence of Hnf4g, gut microfold cell accumulation was substantially impaired relative to that of wildtype mice, as measured by GP2+ cells in small-intestinal Peyer’s patches, a deficit comparable with that of Spib−/− mice, which lack gut microfold cells (Fig. 7 A; Kanaya et al., 2012). We also measured the uptake of fluorescent beads into Peyer’s patches following oral gavage, a well-established assay for microfold-cell activity (Kanaya et al., 2012; Kimura et al., 2019). Hnf4g−/− mice took up fewer fluorescent beads than did controls, indicating a defect in antigen flux across the follicle-associated epithelium (Fig. 7, B and C). The status of thymic Hnf4α did not affect either of these phenotypes.
Figure 7.
Mimetic cells reveal a requirement for Hnf4γ in gut microfold cells. (A) Whole-mount immunofluorescence microscopy of Peyer’s patches from wildtype (left), Spib−/− (center), or Hnf4g−/− mice, stained for the indicated markers. Scale bar, 100 μm. Data are representative of at least two independent experiments. (B and C) Representative microscopy (B) and summarized data (C) of fluorescent-bead (in green) uptake into PPs (nuclei in blue) in mice of the indicated genotypes. Dotted lines indicate follicle-associated epithelium and arrows indicate fluorescent beads. For C, each dot is one PP, bars represent mean ± SEM, data are pooled from three to five mice per genotype and are representative of two independent experiments, and P values were calculated by two-sided, unpaired Student’s t test. Scale bar, 100 μm. (D) Representative flow plots of IgD and IgA expression on B cells from PPs of Hnf4g+/− (top) or Hnf4g−/− (bottom) mice. (E and F) Summarized fraction and number of (E) IgA+ and (F) IgD+ B cells from PPs of Hnf4g+/− (n = 8 for fraction, n = 7 for numbers) and Hnf4g−/− (n = 9) mice. Each dot represents one mouse, bars represent mean ± SEM, data were pooled from two independent experiments, and P values were calculated by two-sided, unpaired Student’s t test. ***, P < 0.001.
Consistent with their loss of microfold cells and antigen flux, Hnf4g−/− mice had impaired adaptive immune responses in the gut, as evidenced by a significant reduction in the proportion and numbers of class-switched IgA+ B cells in the Peyer’s patches of Hnf4g−/− mice relative to controls (Fig. 7, D and E). There was also a reduction in IgD−IgA− B cells, consistent with a general loss of Peyer’s patch germinal center responses in the absence of microfold-cell-driven antigen flux (Fig. 7 D). The defects in gut germinal center responses and IgA production resembled those seen in other models of microfold-cell deficiency, such as Sox8-deficient mice and mice conditionally lacking RANK in the intestinal epithelium (Rios et al., 2016; Kimura et al., 2019). Again, this defect was not affected by the status of thymic Hnf4α (Fig. S5 A). Importantly, the IgA defect did not appear to reflect an intrinsic paucity of B cells themselves as numbers of IgD+ naive B cells in the Peyer’s patches were unchanged (Fig. 7, D and F). Indeed, neither B cells nor any other hematopoietic cell type expressed more than negligible levels of Hnf4g according to the ImmGen RNA-seq atlas (Fig. S5 B).
Figure S5.
The role of Hnf4 in gut microfold cells. (A) Data from Fig. 7 E, stratified by the indicated Hnf4a genotypes. Each dot represents one mouse, bars represent mean ± SEM, and P values were calculated by one-way ANOVA followed by Tukey’s multiple comparisons test. **, P < 0.01. (B) Expression of Hnf4g among all major immunocyte populations, as measured in the ImmGen RNA-seq atlas.
Thus, the requirement for Hnf4 in thymic microfold mTECs revealed an analogous requirement for Hnf4γ in their peripheral counterpart, the gut microfold cell. In Hnf4g−/− mice, gut microfold cell accumulation and function were abnormal, and the gut IgA response was impaired.
Autoimmune characterization of Hnf4ΔTEC mice
Initially, we had planned to analyze the autoimmune consequences of entero-hepato mTEC loss in Hnf4ΔTEC mice. However, our discovery of a role for Hnf4γ in gut microfold cells complicated these analyses, as microfold cells substantially influence systemic immunity (i.e., Kanaya et al., 2012; Kimura et al., 2019). Indeed, we ourselves found that intestinal immunity was altered in the germline absence of Hnf4γ (Fig. 7). Thus, definitive assignment of autoimmunity in Hnf4ΔTEC mice to thymic versus intestinal effects is difficult.
Bearing in mind this caveat, we nonetheless assayed several measures of autoimmunity in these mice. After aging for 15–17 wk, at which age we reliably observe spontaneous autoimmunity in Aire−/− mice, Hnf4ΔTEC mice had lower body weights than control mice, which might have resulted from ongoing inflammation or another cause (Fig. 8 A). However, there was no colonic shortening, splenomegaly, or systemic T cell activation as assessed by splenic T cells (Fig. 8, B–G). We did not observe major spontaneous colitis or hepatitis with aging as assayed by autoantibodies or histological infiltration, but there was a modest increase in liver infiltration in the Hnf4ΔTEC mice (Fig. 8, H and I). Finally, when we challenged these mice in a dextran sodium sulfate (DSS) colitis model, they developed more severe colitis than controls as measured by weight loss (Fig. 8 J).
Figure 8.
Autoimmune characterization of Hnf4ΔTEC mice. (A–C) Hnf4Ctrl and Hnf4ΔTEC mice (littermates, males) were aged 15–17 wk, then assessed for their (A) body weights, (B) colon lengths, and (C) spleen weights (normalized to body weight). (D–G) Activation status of splenic (D) CD4+ Foxp3−, (E) CD4+Foxp3+, and (F and G) CD8+ T cells from mice aged as described in A–C. (H) Autoantibody titers in serum from mice aged as described in A–C compared with serum from sex- and age-matched Aire−/− mice as a positive control. Titers were normalized such that mean signal in serum from Rag1−/− mice was set to zero. (I) Histological disease score of immunocyte infiltration and tissue destruction of the indicated organs in mice aged as described in A–C. Scores ranged from 0 (no disease) to 4 (severe infiltration and tissue destruction). For A–I, each dot represents one mouse, bars show mean ± SEM, data were pooled from three to four independent experiments, and P values were calculated by unpaired, two-sided Student’s t test. (J) Weights of 6-wk-old Hnf4Ctrl (n = 5) and Hnf4ΔTEC (n = 5) mice exposed to 2.5% DSS in the drinking water. Dots show mean weights for each condition, bars show SEM, P value for the difference between genotypes was calculated by two-way ANOVA, and data are representative of two independent experiments. *, P < 0.05; **, P < 0.01.
Altogether, Hnf4ΔTEC mice showed mixed evidence of spontaneous autoimmunity with age and were more susceptible to induced colitis than control mice. Future studies, ideally using an Hnf4g-conditional allele, are warranted to clarify the etiology of these phenotypes.
Discussion
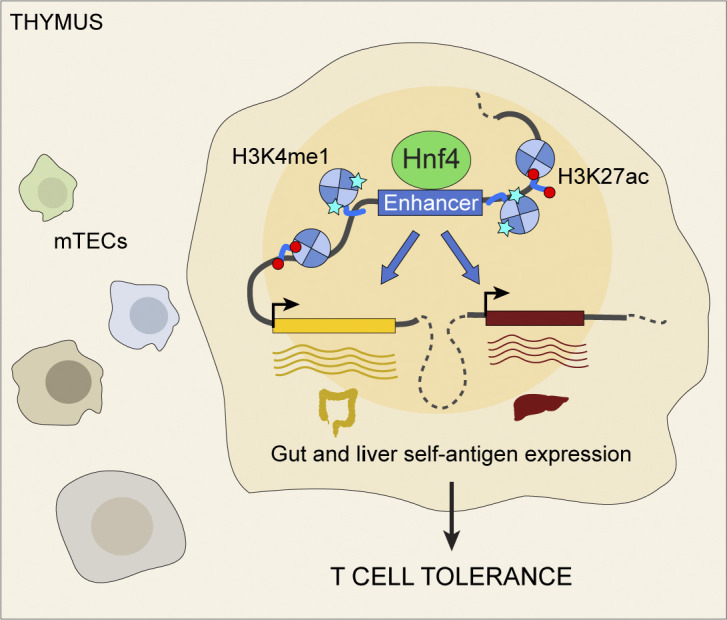
We performed a detailed mechanistic dissection of entero-hepato mTECs as a model for mimetic cell differentiation and function. Entero-hepato mTECs retained a core thymic transcriptional program but layered on a cell-type-specific program broadly resembling that of enterocytes. The entero-hepato mTEC program was driven by binding of Hnf4 to mTEC chromatin and subsequent enhancer activation. Deletion of both Hnf4 family members in thymic epithelium ablated entero-hepato mTECs and their associated PTA expression while prompting a reciprocal increase in secretory mTECs and a transformation of the microfold mTEC transcriptome. The discovery of a role for Hnf4 in microfold mTECs in turn revealed a requirement for Hnf4γ in gut microfold cells.
How, mechanistically, mimetic cells can assume peripheral phenotypes without becoming ectopic or cancerous growths has been puzzling. Our comparison of chromatin and transcriptomes in entero-hepato mTECs and enterocytes showed that mimetic cells do not convert to bona fide peripheral cell types, but rather layer genomic and transcriptomic programs onto a core, retained mTEC identity. Furthermore, the hybrid phenotype of entero-hepato mTECs, with aspects of liver and gut epithelium, emphasizes that mimetic cells are fundamentally TF-driven, blurring lines between peripheral tissues and reflecting the multiple, context-dependent activities of most TFs. Hnf4 was the keystone of the entero-hepato mTEC program, reflecting the critical roles of this family in gut and liver. The centrality of TFs in mimetic cells also explains how the endoderm-derived thymic epithelium can produce mimetic cells that would appear, at first blush, to violate the primacy of germ layers in development, with ectoderm (i.e., skin), mesoderm (i.e., muscle), and endoderm (i.e., gut/liver) all represented.
The loss of entero-hepato mTECs and reciprocal increase in secretory mTECs upon deletion of Hnf4, a phenotype very similar to that observed in the gut upon deletion of Hnf4 (Chen et al., 2019), suggest that mimetic cell differentiation occurs through a series of bi- or multifurcating lineage decisions. Future work tracing mTEC progenitors into different mimetic cell fates should determine whether individual progenitors can give rise to multiple mimetic cell types, what the determinants of such lineage decisions might be, whether different progenitors are restricted in their outputs, and whether interconversion between types is permitted. Indeed, entire medullary islets are known to be clonal, raising interesting questions about what preset biases may exist already at an early stage in mTEC differentiation (Rodewald et al., 2001). We also speculate that the same external cues and morphogens that are so important in peripheral development may influence the differentiation of mimetic cells, a phenomenon already hinted at for RANK signaling and microfold mTECs (Givony et al., 2022 Preprint).
While entero-hepato mTECs were lost in the absence of Hnf4, microfold mTECs were preserved, albeit at a lower frequency and in an altered transcriptomic state. This result was somewhat surprising, given that we had previously postulated that microfold mTECs arise from entero-hepato mTECs based on RNA velocity analysis and lineage progression in the gut (Gebert et al., 1999; Michelson et al., 2022b). We can conceive of two explanations. In one scenario, microfold mTECs may be capable of differentiating from secretory progenitors and, in the absence of entero-hepato mTECs, fill the voided microfold “niche” through this secretory route. Alternatively, microfold mTECs may directly differentiate from Aire-stage mTECs through the combinatorial actions of Hnf4, SpiB, Sox8, and other microfold transcription factors and, while loss of Hnf4 downregulates its target gene program, it does not suffice to collapse microfold cell identity altogether. Lineage analysis, as outlined above, should distinguish between these two models.
Finally, the discovery of a role for Hnf4γ in gut microfold cells based on its analogous function in the thymus shows that we can study the thymus to learn peripheral tissue biology. The thymus has long been described as a “mirror” of the peripheral self (Derbinski et al., 2001), but so far this mirror has been one-way, with the periphery informing our understanding of the thymus but not vice versa. By reflecting back information from the thymus onto the periphery, we can uncover new biology throughout the body.
Materials and methods
Mice
Mouse work was performed at Harvard Medical School following procedures approved by the Harvard Medical School Animal Care and Use Committee, protocol #IS00001257. Strains used were wildtype C57BL/6J (B6; #000664; JAX), Foxn1cre (#018448; JAX; Gordon et al., 2007), Hnf4aflox (Parviz et al., 2002), Hnf4g−/− (Chen et al., 2019), Spib−/− (Sasaki et al., 2012, gift from Dr. Tsuneyasu Kaisho, Wakayama Medical University, Wakyama, Japan), Aire−/− (our colony), and Rag1−/− (#002216; JAX), all maintained on a B6 background. Hnf4aflox and Hnf4g−/− mice were received on a mixed B6, BALB/c, and agouti background, and were backcrossed onto B6 for six generations before use in experiments. Mice were used for experiments between 4 and 8 wk of age, unless otherwise noted. Littermates were used for comparisons of wildtype and knockout mice. Both male and female mice were used for experiments after confirming no apparent difference between sexes. Female mice were used for genomics experiments to control for the effects of sex chromosomes.
Reanalysis of scRNA-seq datasets
scRNA-seq of mimetic cells from Michelson et al. (2022b) was reanalyzed with particular attention to entero-hepato mTECs using the Seurat package (Hao et al., 2021). Briefly, count matrices from mimetic cell scRNA-seq experiments were merged; the gene expression data were jointly normalized; and PCA dimensionality reduction, Louvain clustering, and UMAP visualization were performed on the top 40 principal components. Inspection of the distribution of cells in UMAP space showed good mixing of cells between experiments and no apparent batch effect. Mimetic cells were subset from other mTECs, and dimensionality reduction and visualization were performed a second time on the top 30 principal components. We took a semisupervised clustering approach, performing Louvain clustering of cells (resolution = 1) followed by manual inspection and joining of biologically linked clusters, to assign mimetic cell identities. Expression of key marker genes within each mimetic cell cluster confirmed appropriate assignments.
Isolation, analysis, and sorting of mTECs
mTECs were isolated, analyzed, and sorted as previously described (Michelson et al., 2022a). Thymi were finely chopped, the lymphocyte-rich supernatant was removed, and the thymic pieces were incubated at 37°C in DMEM supplemented with 2% FCS, 25 mM HEPES buffer (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; Lonza), 0.5 mg/ml collagenase (Sigma-Aldrich), and 0.1 mg/ml DNaseI (Sigma-Aldrich) for 15 min, then in the same medium with 0.5 mg/ml collagenase/dispase (Roche) for 15 min. To dissociate cell–cell interactions, 10 mM EDTA was added. To prepurify mTECs, cells were incubated with anti-CD45 microbeads (Miltenyi) for 15 min and then CD45+ cells were depleted using MACS LS columns (Miltenyi). Cells were then stained with antibodies against EpCAM, CD45, Ly51, A/E, Lypd8 (all Biolegend), and/or GP2 (MBL). DAPI (Sigma-Aldrich) and Fixable Yellow Live/Dead (Invitrogen) were used for dead cell exclusion. mTECs were defined as live CD45− EpCAM+ Ly51− cells and further gated as mTEChi or mTEClo based on MHCII levels. Flow cytometry was performed on LSRII or FACSymphony A1 instruments (BD), and cell sorting was performed on a FACSAria cell sorter (BD). Flow cytometry data were analyzed with Flowjo (BD).
Generation of bulk RNA-seq libraries
Bulk RNA-seq libraries were prepared following ImmGen standard operating procedures (https://www.immgen.org). Briefly, 1,000 cells were cytofluorometrically purified into 5 μl TCL buffer (QIAGEN) plus 1% 2-mercaptoethanol (Sigma-Aldrich), and Smartseq2-based bulk RNA-seq libraries were constructed by the Broad Institute Genomics Platform. Entero-hepato mTECs were sorted as Lypd8+GP2− mTECs. Non-entero-hepato mTECs were sorted as Lypd8−GP2− mTECs. Post-Aire mTECs/mimetic cells were sorted as Pdpn−CD104− mTEClo.
Analysis of bulk RNA-seq datasets
Bulk RNA-seq data were preprocessed following ImmGen protocols. Reads were aligned to the mm10 genome using STAR aligner and gene counts were calculated using featureCounts from the Subread package. Samples with fewer than 8,000 genes, high hematopoietic-cell transcript contamination, median transcript integrity number for housekeeping transcripts <45, or poor intrareplicate correlation were excluded from downstream analysis. Downstream analyses were performed in R. DESeq2-normalized expression values (the ImmGen default) were used for direct comparison of gene expression values among samples (Love et al., 2014). For differential expression, edgeR was used to normalize library sizes and calculate fold changes and P values using the quasi-likelihood F-test (Robinson et al., 2010). P values for signature analyses were calculated by one-way Chi-squared test, with the null expectation of no bias in signature up- or downregulation. For comparison between entero-hepato mTECs, other mTECs, enterocytes, and lung alveolar epithelial cells, expression values were jointly normalized across tissues using scaling factors calculated in edgeR. For heatmaps of scaled expression, DESeq2-normalized, log2-transformed expression values were scaled such that the mean expression of each gene was centered on zero but the variation in gene expression was globally scaled. Gene ontology pathway analysis was performed using the online tool gProfiler (https://biit.cs.ut.ee/gprofiler/).
Immunofluorescence microscopy of thymic sections
Imaging of thymic sections was performed as previously described (Michelson et al., 2022a). Briefly, thymi were fixed for 1 h at 4°C in 4% paraformaldehyde (PFA) in PBS and then dehydrated in a 5–30% sucrose gradient overnight. Thymi were embedded in OCT, cut into 8-μm sections, permeabilized and blocked with 5% normal donkey serum in PBS plus 0.05% Tween-20 (PBS-T), stained for 1 h at room temperature (RT) with primary antibodies, washed, stained for 1 h at RT with secondary antibodies, washed, counterstained for nuclei, and mounted for imaging. Primary antibodies used were anti-EpCAM, -Lypd8 (both Biolegend), -GP2 (MBL), -Hnf4α (Abcam), and -Hnf4γ (Proteintech). FITC-, Cy3-, or Cy5-conjugated donkey anti-rat and anti-rabbit secondary antibodies, all from Jackson ImmunoResearch, were used as appropriate. Hoescht 33342 (Sigma-Aldrich) was used as a nuclear counterstain. Images were acquired by widefield microscopy using a Nikon Ti inverted microscope; Plan Apo 10× air, 20× air, or 60× oil objectives; Andor Zyla 4.2 Plus sCMOS camera; and Nikon Elements acquisition software, or by spinning-disk confocal microscopy with the same setup, plus a W1 Yokogawa spinning disk with 50-μm pinholes across multiple z-planes. Images were analyzed in ImageJ.
Reanalysis of scATAC-seq datasets
scATAC-seq of mimetic cells from Michelson et al. (2022b) was reanalyzed with particular attention to entero-hepato mTECs using the SnapATAC package (Fang et al., 2021). Briefly, mimetic cells were subset from other mTECs, dimensionality reduction was performed using a two-step approach involving initial dimensionality reduction via construction of a Jaccard similarity matrix followed by normalization for sequencing depth, and further dimensionality reduction using diffusion maps. UMAP visualization was performed on the top 10 resulting eigenvectors. We again took a semisupervised clustering approach, first performing Louvain clustering (resolution = 1) followed by manual inspection and joining of biologically discrete clusters, to assign mimetic cell identities. Enhanced accessibility of marker gene bodies and enrichment of marker TF motifs within each mimetic cell cluster confirmed appropriate assignments. Motif analysis was performed using chromVAR (Schep et al., 2017).
Generation of CUT&Tag libraries
CUT&Tag libraries for Hnf4γ binding in thymic (n = 4 replicates) and intestinal (n = 2) epithelial cells were generated following a standard approach (Kaya-Okur et al., 2019). Approximately, 100,000 cells were bound to concavalin A beads (Bangs Laboratories), permeabilized in wash buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mM spermidine [Sigma-Aldrich]) plus 0.05% digitonin and incubated overnight at 4°C with 1:50 rabbit polyclonal anti-Hnf4γ (Proteintech). The following morning, some samples were lightly fixed with 0.1% PFA for 2 min at RT. (Note: we and others have observed improved signal for some TFs with light fixation; however, we did not observe differences in thymic Hnf4γ signal in fixed versus unfixed samples, and fixation was omitted for intestinal Hnf4γ libraries). Samples were then incubated with 1:100 secondary guinea pig anti-rabbit IgG (Rockland) for 1 h at RT, washed, incubated with 1:200 pA-Tn5 (#124601; Addgene, purified in-house) for 1 h at RT, washed, tagmented in 300 mM NaCl wash buffer plus 0.01% digitonin for 1 h at 37°C, heat-killed with EDTA, sodium dodecyl sulfate, and proteinase K for 1 h at 55°C, phenol-chloroform extracted, and PCR-amplified using NEBNext 2X Master Mix (NEB) and indexed primers (Buenrostro et al., 2015). The PCR program was 72°C for 2 min, 98°C for 30 s, 16 cycles of 98°C for 10 s and 63°C for 10 s, 72°C for 1 min, hold. Amplified libraries were cleaned and size-selected using AMPure XP beads (Beckman Coulter) and quality-controlled by Qubit (Thermo Fisher Scientific) and Tapestation (Agilent). Intestinal epithelial cells were prepared as previously described (Michelson et al., 2022b). Briefly, small intestines were cleaned, finely chopped, washed extensively, incubated for 1 h in PBS plus 20 mM EDTA, and then shaken vigorously five times, saving each sequential fraction of liberated epithelial cells. Cells were stained with antibodies against EpCAM, CD45, and CD31 (all Biolegend), and epithelial cells were sorted as EpCAM+ CD45− CD31− cells. Purified intestinal epithelial cells were processed for CUT&Tag identically to mTECs.
Analysis of CUT&Tag libraries
Pre-processing, pileup, peak, and motif analyses for CUT&Tag data were performed following published procedures (Michelson et al., 2022b). Briefly, reads were trimmed for adaptors and filtered for quality using Trimmomatic and aligned to the mm10 genome using bowtie2 with the parameters: “--very-sensitive --local --no-mixed --no-discordant -I 10 -X 700” (Langmead and Salzberg, 2012). BAM files were further processed using Samtools and Picard to remove unmapped reads and duplicates, sort reads, and index files (Li et al., 2009). CUT&Tag signals were output as counts per million (CPM)–normalized bigwig files, visualized as genome browser tracks using the Integrative Genomics Viewer, and peak heatmaps generated using deeptools (Ramírez et al., 2016). We used SEACR to call the top 1% of peaks over background under “stringent” mode (Meers et al., 2019). Peaks were used as input to HOMER “findMotifsGenome” routine using default settings to generate de novo motif calls (Heinz et al., 2010). The HOMER routine “annotatePeaks” was used to measure enrichment of different types of genomic elements within peak sets.
Generation of CUT&RUN libraries
mTECs were prepared for CUT&RUN (Skene and Henikoff, 2017) of various chromatin marks by isolating and staining mTECs as above, fixing for 1 min in 0.1% PFA, quenching with 125 mM glycine, cytofluorimetrically sorting 80,000 mTECs per replicate, adding nuclear extraction buffer (final concentrations 20 mM HEPES, 10 mM KCl, 0.1% Triton X-100, 20% glycerol, 1 mM dithiothreitol, 0.5 mM spermidine, 1X Roche complete protease inhibitor) and KDAC inhibitor cocktail (final concentrations 1 μM trichostatin A, 0.5 mM sodium butyrate, 0.5 mM nicotinamide), and slow-freezing samples to −80°C. CUT&RUN library preparation for H3K27ac, H3K4me1, H3K4me3, H3K36me3, H3K27me3, H3.3, CTCF, and IgG was performed for each replicate (i.e., 10,000 cells per factor) via an ImmGen-Epicypher collaboration.
Analysis of CUT&RUN libraries
CUT&RUN data were analyzed similarly to CUT&Tag, with a few modifications. Reads were trimmed for adaptors using TrimGalore and aligned to the mm10 genome using bowtie2 with the parameters: “--very-sensitive --local --no-mixed --no-discordant --phred33 -I 10 -X 1000” (Langmead and Salzberg, 2012). BAM files were further processed using Samtools and Picard to remove unmapped reads and duplicates, sort reads and index files (Li et al., 2009). Bigwig files were generated using deeptools with CPM normalization and were visualized in Integrative Genomics Viewer. Profile plots were generated using deeptools. Entero-hepato–specific and neurosecretory-specific OCRs were generated by calling peaks in scATAC-seq clusters using macs2 (Zhang et al., 2008) and filtering using Bedtools (Quinlan and Hall, 2010) such that only peaks unique to each cluster versus all other clusters were retained.
Generation of scRNA-seq libraries
scRNA-seq libraries of mimetic cells from Hnf4Ctrl and Hnf4ΔTEC mice were generated as previously described (Michelson et al., 2022b). Briefly, mTECs were cytofluorimetrically purified and single-cell libraries generated by the Broad Genomics Platform using droplet-based 3′ V3.1 scRNA-seq chemistry (10X Genomics), following the manufacturer’s instructions. mTECs from three Hnf4Ctrl and four Hnf4ΔTEC mice were pooled, hashing mTECs from each mouse with unique anti-MHCI antibody derived tags to allow for computational demultiplexing.
Analysis of scRNA-seq libraries
Analysis of scRNA-seq of mimetic cells from Hnf4Ctrl and Hnf4ΔTEC mice proceeded largely as described for reanalysis of scRNA-seq of mimetic cells, using Seurat. The top 30 principal components were retained for clustering and visualization. For subclustering of secretory, entero-hepato, and microfold mTECs, these clusters were subset from the main dataset for dimensionality rereduction and revisualization using the top 10 principal components. For differential expression analysis of microfold mTECs from Hnf4Ctrl and Hnf4ΔTEC mice, to avoid the false discoveries associated with cell-level differential expression analysis (Squair et al., 2021), pseudobulk counts were calculated for each gene on a per-replicate basis, and these pseudobulk counts were used as inputs to edgeR to perform differential expression analysis as outlined for bulk RNA-seq.
Whole-mount imaging of Peyer’s patches (PPs)
Whole-mount imaging of PPs was performed following a published protocol (Lai et al., 2020), with some modifications. Briefly, PPs were isolated from mouse small intestines, trimmed, and vortexed vigorously for 30 s, five times, in 1 ml PBS plus 5% FCS, 1 μM nicardipine, 5 mM dithiothreitol, and 0.05% Tween-20 to remove mucus and relax smooth muscle. PPs were then fixed flat in 4% PFA for 1 h at 4°C, blocked and permeabilized in PBS plus 5% rat serum and 0.5% Triton X-100 for 1 h at 4°C, stained overnight in PBS plus 5% rat serum, 0.1% Tween-20 and 1:1,000 anti-GP2 (MBL), washed five times in PBS plus 0.1% Tween-20, counterstained with 1:1,000 Hoescht 33342 (Sigma-Aldrich) for 15 min, washed again, and mounted in Prolong Diamond antifade mountant (Invitrogen) with the luminal side facing the coverslip. PPs were imaged by spinning-disk confocal microscopy across multiple z-planes using a Nikon Ti inverted microscope, W1 Yokogawa spinning disk with 50 μm pinholes, Plan Apo 10× air objective, Andor Zyla 4.2 Plus sCMOS camera, and Nikon Elements acquisition software.
Measurement of fluorescent bead uptake into PPs
Uptake of fluorescent beads into PPs was measured following published protocols (Kanaya et al., 2012; Kimura et al., 2019). Briefly, mice were fasted overnight, then orally gavaged with 2*1012 200 nm Fluoresbrite yellow–green fluorescent beads (Polysciences). 3 h later, PPs were isolated, flash-frozen, sectioned, and nuclei-counterstained with Hoescht. PPs were imaged by widefield microscopy as described for immunofluorescence using a 20× air objective. Analysis was performed by tracing PP areas in ImageJ and manually counting the number of fluorescent beads within them.
Flow cytometry of hematopoietic cells
PP B cells were analyzed by isolating PPs, passing them over a 70-μm filter, staining a small fraction of cells with antibodies against CD45, CD19, B220, IgD, CD11b, CD138 (all Biolegend), IgA, and CD11c (both eBioscience), and excluding dead cells using Fixable Yellow Live/Dead (Invitrogen). B cells were gated as live CD45+CD19+B220+ cells. Cell numbers were normalized using 123count eBeads (Invitrogen). Thymi and spleens for T cell analysis were prepared similarly, using antibodies against CD45, TCRβ, CD8α, CD25, CD44, CD62L, CD73 (all Biolegend), CD4, and Foxp3 (both eBioscience).
Serum autoantibody and tissue histology analysis
Hnf4Ctrl and Hnf4ΔTEC mice (males, littermates) were cohoused and aged to 15–17 wk. Mice were euthanized, serum was obtained for autoantibody analysis, and eyes, liver, and colon were taken for histological analysis. For autoantibody analysis, serum from each mouse was diluted 1:100 in PBS-T plus 5% donkey serum and incubated on liver and colon sections from Rag1−/− mice for 1 h at RT. Sections were then washed and incubated with Cy5 donkey anti-mouse IgG (Jackson ImmunoResearch) for 1 h at RT. Sections were washed, counterstained with Hoescht, and imaged by widefield immunofluorescence microscopy using the previously described microscope. Autoantibody signal was quantified for each image in ImageJ by selecting identically sized regions of interest, calculating their integrated density, and subtracting the background signal from a section stained with Rag1−/− serum. For histological analysis, organs were fixed in 10% formalin solution, embedded, sectioned, stained with hematoxylin and eosin by the Harvard Medical School Rodent Histopathology Core, and scored by a trained, blinded evaluator. For eye and liver, lymphocyte infiltration and tissue destruction were scored from 0 (none) to 4 (severe). For colon, each colon was scored from 0 (none) to 4 (severe) for lymphocytic infiltration, mucosal ulceration, and goblet cell loss independently, and the overall score was calculated as an average of the three.
DSS colitis
6-wk-old Hnf4Ctrl and Hnf4ΔTEC littermate mice were treated with 2.5% DSS for 6 d in their drinking water followed by 4 d of recovery. Body weight was measured every 1–2 d in a blinded fashion.
Statistical analysis
P values for de novo motif enrichment were calculated in HOMER by binomial test. P values for scRNA-seq differential expression were calculated in Seurat using the Wilcoxon rank sum test. P values for bulk RNA-seq differential expression were calculated in edgeR using the quasi-likelihood F-test. P values for enrichment of gene signatures in bulk RNA-seq data were calculated by one-way chi-squared test. P values for flow cytometry data were calculated by unpaired, two-sided Student’s t test. P values were adjusted for multiple comparisons by the Benjamini-Hochberg (BH) method where indicated in the figure legends. Sample sizes and other statistical tests are indicated in the figure legends. P = *, <0.05; **, <0.01; ***, <0.001; ****, <0.0001. Statistical analyses were performed using R (v4.0.2) or GraphPad Prism (v7).
Online supplemental material
Fig. S1 shows marker genes and motifs for mimetic-cell scRNA-seq and scATAC-seq clusters as well as mTEC gating strategy and additional characterization of entero-hepato mTECs. Fig. S2 quantitates thymocyte and TEC subsets in Hnf4Ctrl and Hnf4ΔTEC mice. Fig. S3 displays additional characterization of the chromatin landscape of mTECs from Hnf4Ctrl and Hnf4ΔTEC mice. Fig. S4 shows marker genes and mimetic cell frequencies in scRNA-seq of mimetic cells from Hnf4Ctrl and Hnf4ΔTEC mice. Fig. S5 displays additional characterization of Hnf4γ’s role in gut microfold cells. Table S1 contains metadata and quality control metrics for CUT&Tag and CUT&RUN data.
Supplementary Material
contains metadata and quality control metrics for CUT&Tag and CUT&RUN data.
Acknowledgments
We gratefully acknowledge Ian Magill for generating scRNA-seq libraries; Juliana Lee for generating population-level RNA-seq and CUT&RUN libraries; Kimie Hattori and Adriana Ortiz-Lopez for animal husbandry and general experimental assistance; Liang Yang, Niket Patel, and Dania Mallah for pre-processing of RNA-seq data; Catherine Laplace for figure preparation; Soumya Raychaudhuri and Ruth Franklin for helpful discussions; and the Broad Genomics Platform, Harvard Biopolymers Sequencing Core, Harvard Immunology Flow Core, and Harvard Rodent Histopathology Core for experimental support.
This work was funded by National Institutes of Health grants R01AI088204 and R01DK060027 (to D. Mathis), T32GM007753 and T32GM144273 (for D.A. Michelson), and R01DK126446 and R01DK121915 (to M. Verzi).
Author contributions: D.A. Michelson and D. Mathis conceived the study. D.A. Michelson performed most of the experiments and analyzed the data. C. Zuo performed and analyzed the bead uptake and DSS colitis assays. M. Verzi provided the Hnf4aflox Hnf4gKO mice. D. Mathis and C. Benoist supervised the work. D.A. Michelson and D. Mathis wrote the manuscript, which all authors critically reviewed. D. Mathis acquired funding.
Data availability
CUT&Tag, CUT&RUN, population-level RNA-seq, and scRNA-seq data from this paper are available at GEO (GSE225661). Scripts used to analyze sequencing data are available at GitHub (http://github.com/dmichelson). Requests for any other data should be directed to and will be fulfilled by the corresponding author.
References
- Abramson, J., Giraud M., Benoist C., and Mathis D.. 2010. Aire’s partners in the molecular control of immunological tolerance. Cell. 140:123–135. 10.1016/j.cell.2009.12.030 [DOI] [PubMed] [Google Scholar]
- Anderson, M.S., Venanzi E.S., Klein L., Chen Z., Berzins S.P., Turley S.J., von Boehmer H., Bronson R., Dierich A., Benoist C., and Mathis D.. 2002. Projection of an immunological self shadow within the thymus by the aire protein. Science. 298:1395–1401. 10.1126/science.1075958 [DOI] [PubMed] [Google Scholar]
- Apostolou, I., Sarukhan A., Klein L., and von Boehmer H.. 2002. Origin of regulatory T cells with known specificity for antigen. Nat. Immunol. 3:756–763. 10.1038/ni816 [DOI] [PubMed] [Google Scholar]
- Bansal, K., Michelson D.A., Ramirez R.N., Viny A.D., Levine R.L., Benoist C., and Mathis D.. 2021. Aire regulates chromatin looping by evicting CTCF from domain boundaries and favoring accumulation of cohesin on superenhancers. Proc. Natl. Acad. Sci. USA. 118:e2110991118. 10.1073/pnas.2110991118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal, K., Yoshida H., Benoist C., and Mathis D.. 2017. The transcriptional regulator Aire binds to and activates super-enhancers. Nat. Immunol. 18:263–273. 10.1038/ni.3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensinger, S.J., Bandeira A., Jordan M.S., Caton A.J., and Laufer T.M.. 2001. Major histocompatibility complex class II-positive cortical epithelium mediates the selection of CD4(+)25(+) immunoregulatory T cells. J. Exp. Med. 194:427–438. 10.1084/jem.194.4.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland, P.W., and Warren L.G.. 1986. Antigen presentation by epithelial cells of the rat small intestine. I. Kinetics, antigen specificity and blocking by anti-Ia antisera. Immunology. 58:1–7. [PMC free article] [PubMed] [Google Scholar]
- Bornstein, C., Nevo S., Giladi A., Kadouri N., Pouzolles M., Gerbe F., David E., Machado A., Chuprin A., Tóth B., et al. 2018. Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature. 559:622–626. 10.1038/s41586-018-0346-1 [DOI] [PubMed] [Google Scholar]
- Brennecke, P., Reyes A., Pinto S., Rattay K., Nguyen M., Küchler R., Huber W., Kyewski B., and Steinmetz L.M.. 2015. Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat. Immunol. 16:933–941. 10.1038/ni.3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro, J.D., Wu B., Litzenburger U.M., Ruff D., Gonzales M.L., Snyder M.P., Chang H.Y., and Greenleaf W.J.. 2015. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 523:486–490. 10.1038/nature14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Toke N.H., Luo S., Vasoya R.P., Fullem R.L., Parthasarathy A., Perekatt A.O., and Verzi M.P.. 2019. A reinforcing HNF4-SMAD4 feed-forward module stabilizes enterocyte identity. Nat. Genet. 51:777–785. 10.1038/s41588-019-0384-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbinski, J., Pinto S., Rösch S., Hexel K., and Kyewski B.. 2008. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc. Natl. Acad. Sci. USA. 105:657–662. 10.1073/pnas.0707486105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbinski, J., Schulte A., Kyewski B., and Klein L.. 2001. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat. Immunol. 2:1032–1039. 10.1038/ni723 [DOI] [PubMed] [Google Scholar]
- DeVoss, J., Hou Y., Johannes K., Lu W., Liou G.I., Rinn J., Chang H., Caspi R.R., Fong L., and Anderson M.S.. 2006. Spontaneous autoimmunity prevented by thymic expression of a single self-antigen. J. Exp. Med. 203:2727–2735. 10.1084/jem.20061864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalla, F., Baran-Gale J., Maio S., Chappell L., Holländer G.A., and Ponting C.P.. 2020. Biologically indeterminate yet ordered promiscuous gene expression in single medullary thymic epithelial cells. EMBO J. 39:e101828. 10.15252/embj.2019101828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley, J., Erickson M., and Farr A.G.. 2005. An organized medullary epithelial structure in the normal thymus expresses molecules of respiratory epithelium and resembles the epithelial thymic rudiment of nude mice. J. Immunol. 175:4331–4337. 10.4049/jimmunol.175.7.4331 [DOI] [PubMed] [Google Scholar]
- Fang, R., Preissl S., Li Y., Hou X., Lucero J., Wang X., Motamedi A., Shiau A.K., Zhou X., Xie F., et al. 2021. Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat. Commun. 12:1337. 10.1038/s41467-021-21583-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr, A.G., Dooley J.L., and Erickson M.. 2002. Organization of thymic medullary epithelial heterogeneity: Implications for mechanisms of epithelial differentiation. Immunol. Rev. 189:20–27. 10.1034/j.1600-065X.2002.18903.x [DOI] [PubMed] [Google Scholar]
- Gavanescu, I., Kessler B., Ploegh H., Benoist C., and Mathis D.. 2007. Loss of Aire-dependent thymic expression of a peripheral tissue antigen renders it a target of autoimmunity. Proc. Natl. Acad. Sci. USA. 104:4583–4587. 10.1073/pnas.0700259104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert, A., Fassbender S., Werner K., and Weissferdt A.. 1999. The development of M cells in Peyer’s patches is restricted to specialized dome-associated crypts. Am. J. Pathol. 154:1573–1582. 10.1016/S0002-9440(10)65410-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdin, A.K., Surve V.V., Jönsson M., Bjursell M., Björkman M., Edenro A., Schuelke M., Saad A., Bjurström S., Lundgren E.J., et al. 2006. Phenotypic screening of hepatocyte nuclear factor (HNF) 4-γ receptor knockout mice. Biochem. Biophys. Res. Commun. 349:825–832. 10.1016/j.bbrc.2006.08.103 [DOI] [PubMed] [Google Scholar]
- Giraud, M., Yoshida H., Abramson J., Rahl P.B., Young R.A., Mathis D., and Benoist C.. 2012. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc. Natl. Acad. Sci. USA. 109:535–540. 10.1073/pnas.1119351109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givony, T., del Castillo D., Nevo S., Leshkowitz D., Kadouri N., Dassa B., Ben-Nun O., Gruper Y., Gome T., Dobes J., et al. 2022. Thymic microfold and endocrine cells regulate thymus homeostasis and self-tolerance. Res. Square. (Preprint posted July 14, 2022). https://assets.researchsquare.com/files/rs-1837610/v1_covered.pdf?c=1657825917. [Google Scholar]
- Gordon, J., Xiao S., Hughes B. III, Su D.M., Navarre S.P., Condie B.G., and Manley N.R.. 2007. Specific expression of lacZ and cre recombinase in fetal thymic epithelial cells by multiplex gene targeting at the Foxn1 locus. BMC Dev. Biol. 7:69. 10.1186/1471-213X-7-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha, M., Saare M., Maslovskaja J., Kisand K., Liiv I., Haljasorg U., Tasa T., Metspalu A., Milani L., and Peterson P.. 2017. DNA breaks and chromatin structural changes enhance the transcription of autoimmune regulator target genes. J. Biol. Chem. 292:6542–6554. 10.1074/jbc.M116.764704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Y., Hao S., Andersen-Nissen E., Mauck W.M. III, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. 2021. Integrated analysis of multimodal single-cell data. Cell. 184:3573–3587.e29. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassall, A.H. 1846. The Microscopic Anatomy of the Human Body: In Health and Disease. S. Highley, London. [Google Scholar]
- Heinz, S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, M.S., Boesteanu A., Reed A.J., Petrone A.L., Holenbeck A.E., Lerman M.A., Naji A., and Caton A.J.. 2001. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat. Immunol. 2:301–306. 10.1038/86302 [DOI] [PubMed] [Google Scholar]
- Kanaya, T., Hase K., Takahashi D., Fukuda S., Hoshino K., Sasaki I., Hemmi H., Knoop K.A., Kumar N., Sato M., et al. 2012. The Ets transcription factor Spi-B is essential for the differentiation of intestinal microfold cells. Nat. Immunol. 13:729–736. 10.1038/ni.2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappler, J.W., Roehm N., and Marrack P.. 1987. T cell tolerance by clonal elimination in the thymus. Cell. 49:273–280. 10.1016/0092-8674(87)90568-X [DOI] [PubMed] [Google Scholar]
- Kaya-Okur, H.S., Wu S.J., Codomo C.A., Pledger E.S., Bryson T.D., Henikoff J.G., Ahmad K., and Henikoff S.. 2019. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10:1930. 10.1038/s41467-019-09982-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, S., Kobayashi N., Nakamura Y., Kanaya T., Takahashi D., Fujiki R., Mutoh M., Obata Y., Iwanaga T., Nakagawa T., et al. 2019. Sox8 is essential for M cell maturation to accelerate IgA response at the early stage after weaning in mice. J. Exp. Med. 216:831–846. 10.1084/jem.20181604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisielow, P., Blüthmann H., Staerz U.D., Steinmetz M., and von Boehmer H.. 1988. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 333:742–746. 10.1038/333742a0 [DOI] [PubMed] [Google Scholar]
- Klein, L., Klein T., Rüther U., and Kyewski B.. 1998. CD4 T cell tolerance to human C-reactive protein, an inducible serum protein, is mediated by medullary thymic epithelium. J. Exp. Med. 188:5–16. 10.1084/jem.188.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, L., Kyewski B., Allen P.M., and Hogquist K.A.. 2014. Positive and negative selection of the T cell repertoire: What thymocytes see (and don’t see). Nat. Rev. Immunol. 14:377–391. 10.1038/nri3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, N.Y., Musser M.A., Pinho-Ribeiro F.A., Baral P., Jacobson A., Ma P., Potts D.E., Chen Z., Paik D., Soualhi S., et al. 2020. Gut-innervating nociceptor neurons regulate Peyer’s patch microfold cells and SFB levels to mediate Salmonella host defense. Cell. 180:33–49.e22. 10.1016/j.cell.2019.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabbott, N.A., Donaldson D.S., Ohno H., Williams I.R., and Mahajan A.. 2013. Microfold (M) cells: Important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 6:666–677. 10.1038/mi.2013.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers, M.P., Tenenbaum D., and Henikoff S.. 2019. Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin. 12:42. 10.1186/s13072-019-0287-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith, M., Zemmour D., Mathis D., and Benoist C.. 2015. Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat. Immunol. 16:942–949. 10.1038/ni.3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson, D.A., Benoist C., and Mathis D.. 2022a. CTLA-4 on thymic epithelial cells complements Aire for T cell central tolerance. Proc. Natl. Acad. Sci. USA. 119:e2215474119. 10.1073/pnas.2215474119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson, D.A., Hase K., Kaisho T., Benoist C., and Mathis D.. 2022b. Thymic epithelial cells co-opt lineage-defining transcription factors to eliminate autoreactive T cells. Cell. 185:2542–2558.e18. 10.1016/j.cell.2022.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson, D.A., and Mathis D.. 2022. Thymic mimetic cells: Tolerogenic masqueraders. Trends Immunol. 43:782–791. 10.1016/j.it.2022.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, C.N., Proekt I., von Moltke J., Wells K.L., Rajpurkar A.R., Wang H., Rattay K., Khan I.S., Metzger T.C., Pollack J.L., et al. 2018. Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature. 559:627–631. 10.1038/s41586-018-0345-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura, R., Kurakawa T., Nakano T., Kayama H., Kinoshita M., Motooka D., Gotoh K., Kimura T., Kamiyama N., Kusu T., et al. 2016. Lypd8 promotes the segregation of flagellated microbiota and colonic epithelia. Nature. 532:117–121. 10.1038/nature17406 [DOI] [PubMed] [Google Scholar]
- Oven, I., Brdicková N., Kohoutek J., Vaupotic T., Narat M., and Peterlin B.M.. 2007. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol. Cell. Biol. 27:8815–8823. 10.1128/MCB.01085-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parviz, F., Li J., Kaestner K.H., and Duncan S.A.. 2002. Generation of a conditionally null allele of hnf4alpha. Genesis. 32:130–133. 10.1002/gene.10058 [DOI] [PubMed] [Google Scholar]
- Parviz, F., Matullo C., Garrison W.D., Savatski L., Adamson J.W., Ning G., Kaestner K.H., Rossi J.M., Zaret K.S., and Duncan S.A.. 2003. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat. Genet. 34:292–296. 10.1038/ng1175 [DOI] [PubMed] [Google Scholar]
- Quinlan, A.R., and Hall I.M.. 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez, F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., and Manke T.. 2016. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44:W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remak, R. 1855. Untersuchungen über die Entwickelung der Wirbelthiere. G. Reimer, London. [Google Scholar]
- Rios, D., Wood M.B., Li J., Chassaing B., Gewirtz A.T., and Williams I.R.. 2016. Antigen sampling by intestinal M cells is the principal pathway initiating mucosal IgA production to commensal enteric bacteria. Mucosal Immunol. 9:907–916. 10.1038/mi.2015.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodewald, H.R., Paul S., Haller C., Bluethmann H., and Blum C.. 2001. Thymus medulla consisting of epithelial islets each derived from a single progenitor. Nature. 414:763–768. 10.1038/414763a [DOI] [PubMed] [Google Scholar]
- Rossi, S.W., Kim M.Y., Leibbrandt A., Parnell S.M., Jenkinson W.E., Glanville S.H., McConnell F.M., Scott H.S., Penninger J.M., Jenkinson E.J., et al. 2007. RANK signals from CD4(+)3(−) inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J. Exp. Med. 204:1267–1272. 10.1084/jem.20062497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, I., Hoshino K., Sugiyama T., Yamazaki C., Yano T., Iizuka A., Hemmi H., Tanaka T., Saito M., Sugiyama M., et al. 2012. Spi-B is critical for plasmacytoid dendritic cell function and development. Blood. 120:4733–4743. 10.1182/blood-2012-06-436527 [DOI] [PubMed] [Google Scholar]
- Schep, A.N., Wu B., Buenrostro J.D., and Greenleaf W.J.. 2017. chromVAR: Inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods. 14:975–978. 10.1038/nmeth.4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene, P.J., and Henikoff S.. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife. 6:e21856. 10.7554/eLife.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squair, J.W., Gautier M., Kathe C., Anderson M.A., James N.D., Hutson T.H., Hudelle R., Qaiser T., Matson K.J.E., Barraud Q., et al. 2021. Confronting false discoveries in single-cell differential expression. Nat. Commun. 12:5692. 10.1038/s41467-021-25960-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius, M., Zheng S., Houck-Loomis B., Hao S., Yeung B.Z., Mauck W.M. III, Smibert P., and Satija R.. 2018. Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19:224. 10.1186/s13059-018-1603-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, R.T., DeVoss J.J., Moon J.J., Sidney J., Sette A., Jenkins M.K., and Anderson M.S.. 2012. Detection of an autoreactive T-cell population within the polyclonal repertoire that undergoes distinct autoimmune regulator (Aire)-mediated selection. Proc. Natl. Acad. Sci. USA. 109:7847–7852. 10.1073/pnas.1120607109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, W., Wei Y., Ye Z., Wang J., Yang W., Yu G., Xiong J., and Jia S.. 2023. Insm1 regulates the development of mTECs and immune tolerance. bioRxiv. (Preprint posted January 14, 2023). 10.1101/2023.01.14.524041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaseñor, J., Besse W., Benoist C., and Mathis D.. 2008. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: Probabilistic, monoallelic, misinitiated. Proc. Natl. Acad. Sci. USA. 105:15854–15859. 10.1073/pnas.0808069105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, K.L., Miller C.N., Gschwind A.R., Wei W., Phipps J.D., Anderson M.S., and Steinmetz L.M.. 2020. Combined transient ablation and single-cell RNA-sequencing reveals the development of medullary thymic epithelial cells. Elife. 9:e60188. 10.7554/eLife.60188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, H., Bansal K., Schaefer U., Chapman T., Rioja I., Proekt I., Anderson M.S., Prinjha R.K., Tarakhovsky A., Benoist C., and Mathis D.. 2015. Brd4 bridges the transcriptional regulators, Aire and P-TEFb, to promote elongation of peripheral-tissue antigen transcripts in thymic stromal cells. Proc. Natl. Acad. Sci. USA. 112:E4448–E4457. 10.1073/pnas.1512081112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains metadata and quality control metrics for CUT&Tag and CUT&RUN data.
Data Availability Statement
CUT&Tag, CUT&RUN, population-level RNA-seq, and scRNA-seq data from this paper are available at GEO (GSE225661). Scripts used to analyze sequencing data are available at GitHub (http://github.com/dmichelson). Requests for any other data should be directed to and will be fulfilled by the corresponding author.