Figure 2.
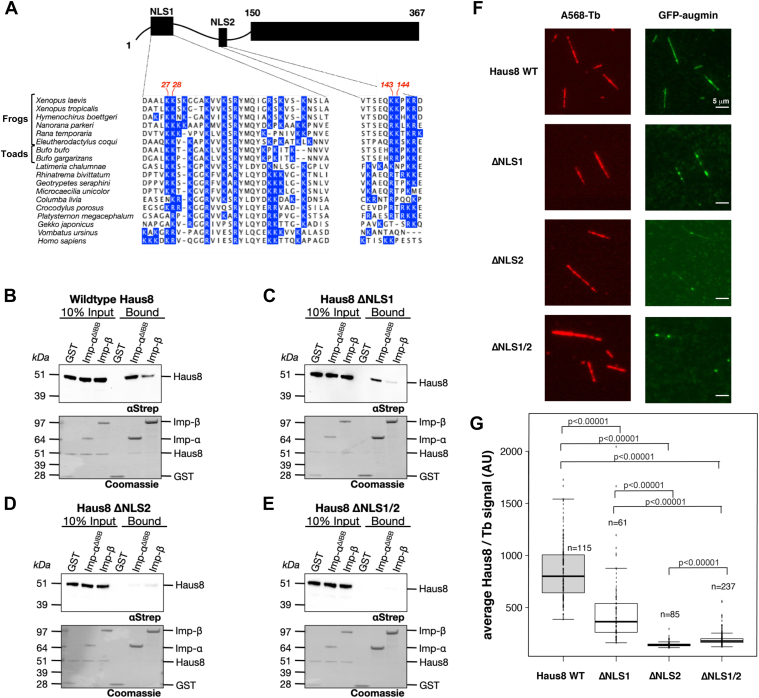
Haus8 of augmin binds to importins and MTs through two NLS sites.A, the augmin subunit Haus8 is predicted to contain two NLS sequences within its disordered N terminus (43). The domain architecture of Haus8 is cartooned at top, and the sequence of each predicted NLS in Xenopus laevis is shown at the bottom. X. laevis Haus8 is shown aligned to other vertebrate orthologs below, and all basic residues (arginine abbreviated as R and lysine as K) are highlighted in blue. Indicated in red at the top of the sequence are the pairs of basic residues mutated to alanine to generate the Haus8 mutants ΔNLS1 (K27A/K28A) and ΔNLS2 (K143A/K144A). B–E, pulldowns of Strep-GFP-Haus81–150, either wildtype or containing the indicated NLS mutants, were conducted as for Figure 1D. F, selected TIRF images of in vitro binding of GST-GFP-Haus81–150 to stabilized MT seeds. Haus8 constructs with mutated residues in NLS1 and/or NLS2 result in a reduction in binding, as quantified in G. Images belonging to the same experiment were contrast matched. To compare augmin fluorescence intensity across experiments, the intensity was normalized with respect to the tubulin signal. G, boxplot of average GFP-Haus8 signal relative to the average tubulin signal, where each marker represents a single MT from the experiment shown in F. The total number of MTs (n) was collected from two replicates. Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. p Values were calculated from independent t tests. GST, glutathione-S-transferase; MT, microtubule; NLS, nuclear localization signal; TIRF, total internal reflection fluorescence.