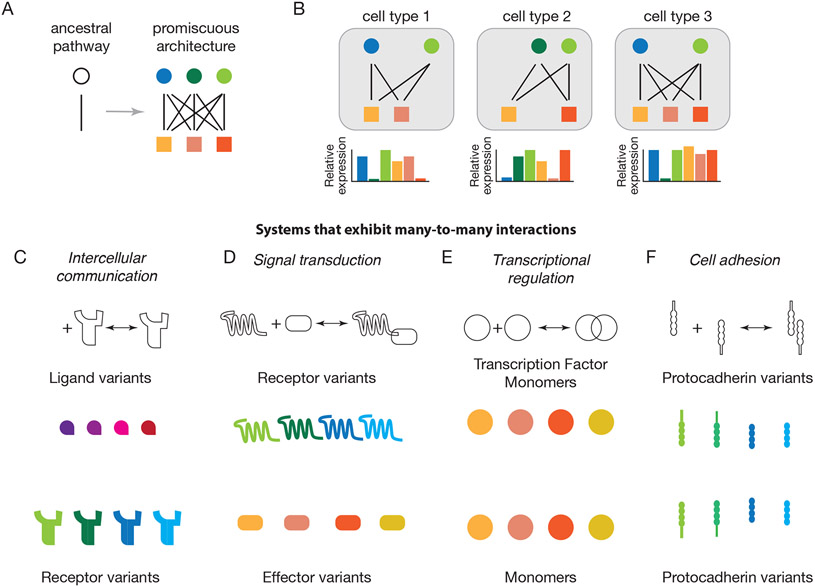
Figure 1: Diverse cellular pathways exhibit many-to-many interactions among protein families.
(A) Extant pathways, such as signaling from one component (circle) to another (square) can evolve from simpler ancestral pathways through gene duplications, resulting in many-to-many interaction networks (right).
(B) Different cell types (gray) typically express different protein variant profiles (schematic bar plots).
(C) Intercellular communication systems often comprise multiple ligand (upper) and receptor (lower) variants that interact in a many-to-many fashion, with each ligand binding to multiple receptors and each receptor binding to multiple ligand variants.
(D) In signal transduction systems, receptor variants (upper) interact with variant intracellular signal transducers, or effectors, in a many-to-many fashion.
(E) In cell adhesion processes, protocadherin variants interact with other protocadherins in adjacent cells, also in a many-to-many fashion.
(F) Eukaryotic transcription factor variants can often bind to one another to form a large repertoire of distinct dimers, each with distinct DNA binding specificities.