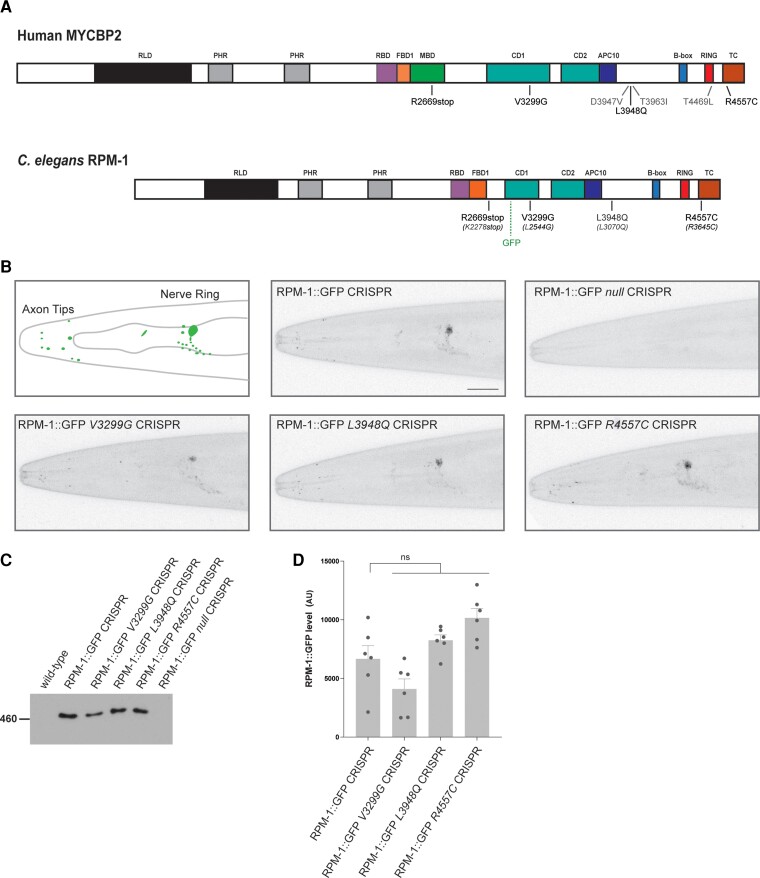
Figure 2.
CRISPR editing MDCD-related variants into rpm-1 does not impair protein localization or stability in C. elegans. (A) Schematic of human MYCBP2 and its C. elegans orthologue RPM-1. Shown are protein sequence changes caused by MYCBP2 missense and nonsense variants from MDCD patients. Also noted is the location of the GFP inserted using CRISPR/Cas9 gene engineering in C. elegans and annotated protein domains. RCC1 like domain (RLD), Pam/Highwire/RPM-1 (PHR) domain, RAE-1 binding domain (RBD), FSN-1 binding domain (FBD), conserved domain of unknown function (CD), anaphase-promoting complex subunit 10 like domain (APC10). (B) CRISPR engineering was used to fuse GFP with endogenous RPM-1. CRISPR editing then introduced conserved MDCD-related variants (V3299G, L3948Q and R4557C) or a known null mutation (C4587Y) into RPM-1::GFP CRISPR animals. Shown is a schematic of RPM-1::GFP CRISPR localization at axon tips and the nerve ring in the head of C. elegans (left). Representative confocal images are shown for all genotypes. Note localization and levels of RPM-1::GFP are normal in CRISPR edited animals carrying MDCD-related variants (V3299G, L3948Q and R4557C). (C) Immunoprecipitation of RPM-1::GFP from whole C. elegans lysates for indicated genotypes. Representative of three independent experiments. (D) Quantitation of RPM-1::GFP for indicated genotypes. Means are shown for six replicates from three independent experiments for each genotype. Error bars represent SEM. Significance determined using Student’s t-test with Bonferroni correction. ns = not significant. Scale bar = 20 μm.