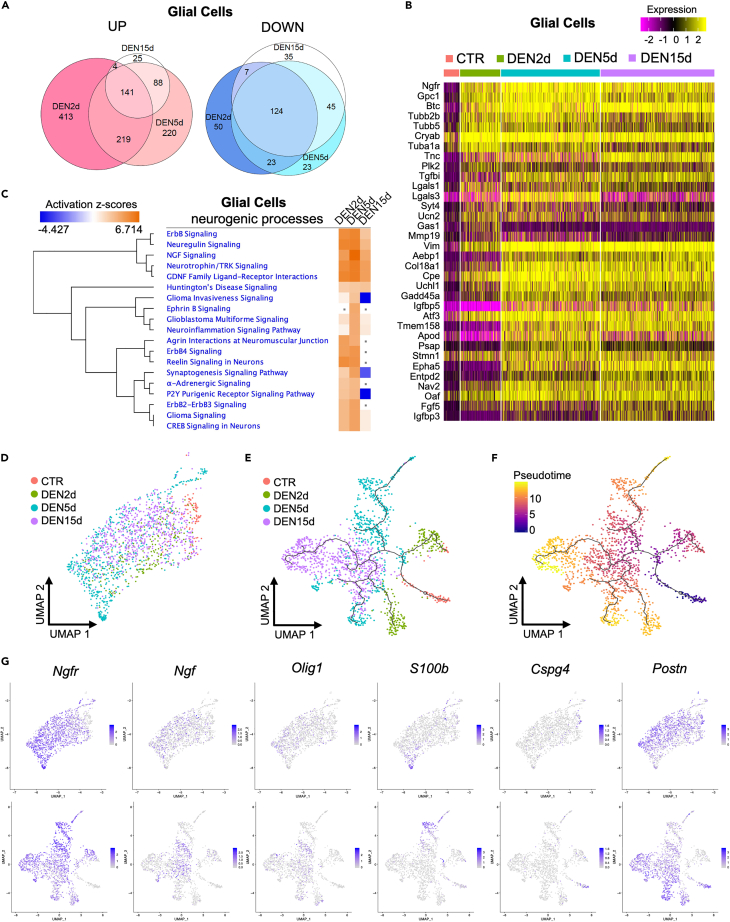
Figure 3.
Response of muscle-resident glial cells to denervation
(A) Venn diagram of upregulated genes (left) and downregulated genes (right) in glial cells at DEN2d, DEN5d, and DEN15d compared with CTR. Genes with p-value-adj ≤ 0.01 are counted.
(B) Heatmap of top 20 upregulated genes in glial cells at DEN2d, DEN5d, and DEN15d respectively compared with CTR. Gene expression values visualized on a scale from low (purple) to high (yellow), with black for zero expression value.
(C) IPA comparison of DE genes in glial cells at DEN2d, DEN5d, and DEN15d respectively compared with CTR. Selected pathways related to neurogenic processes are shown. Pathways with p value ≥ 0.05 were indicated with gray dots. Pathways activation status (Z-scores) visualized on a scale from repressed (blue) to activated (orange), with neutral status (Z-Score = 0) set to white.
(D) UMAP embedding subset from the whole dataset of glial cells colored by time point.
(E) UMAP embedding of glial cells after re-clustering, with superimposition of pseudo-time trajectories from Monocle3. Glial cells are colored by time point.
(F) Same as (E), but glial cells are colored by pseudo-time. Pseudotime visualized on a scale from early (blue) to late (yellow), with intermediate stages in purple and orange.
(G) UMAP embedding of glial cells before (upper) and after (bottom) re-clustering, showing the expression of selected genes associated with different glial cell-derived lineages.